- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
MECHANISM OF PROTEIN SYNTHESIS
The process of protein synthesis consists of two major steps:
(A) Transcription or synthesis of mRNA on DNA
(B) Translation or synthesis of proteins along mRNA
(A) Transcription
The transfer of genetic information from DNA to mRNA is known as transcription. The segment of DNA that takes part in transcription is called transcription unit. It has three components.
(i) A Promoter
(ii) The Structural gene
(iii) A Terminator
Promoter seguences are present upstream (5' end) of the structural genes of a transcription unit.
The binding sites for RNA polymerase lie within the promoter sequence.
Certain short sequences within the promoter sites are conserved. In prokaryotes, 10 bp upstream from, the start point lies a conserved sequence described as –10 sequence TATAAT or "Pribnow box" and –35 sequence TTGACA as "Recognition sequence".
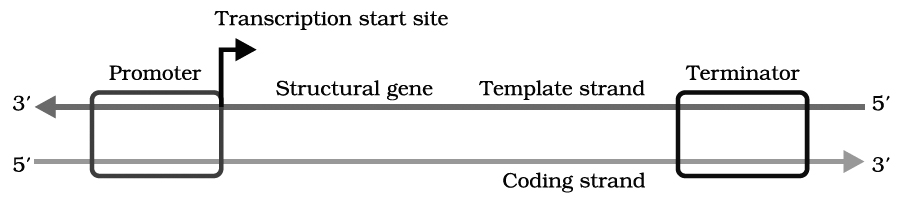
Schematic structure of a transcription unit
Structural gene is part of that DNA strand which has 3' 5' polarity, as transcription occur in 5' 3' direction. This strand of DNA that direct the synthesis of mRNA is called template strand. The complementary strand is called non-template or coding strand, it is identical in base sequence to RNA transcribed from the gene, only with U in place of T.
Terminator is present at 3' end of coding strand and defines the end of the process of transcription.
Mechanism of transcription
RNA polymerase binds to promoter region of the DNA and the process of transcription begins.
RNA polymerase moves along DNA helix and unwinds it.
One of the two strands of DNA serves as a template for RNA synthesis.
This results in the formation of complementary RNA strand.
It is formed at a rate of about 40 to 50 nucleotides per second.
RNA synthesis comes to a stop when RNA polymerase reaches the terminator sequence.
The transcription enzyme, i.e., RNA polymerase is only of one type in prokaryotes and can transcribe all types of RNAs.
RNA polymerase is a holoenzyme that is represented as (2').
Molecular weight of holoenzyme is 4,50,000.
The enzyme without subunit is referred to as core enzyme.
Though, the core enzyme is capable of transcribing DNA into RNA but transcription starts non specifically at any base on DNA.
It is subunit which confers specificity. Rho factor () is required for termination of transcription.
(a) Sigma Factor () : Binds to the promoter site of DNA and it initiates transcription.
(b) Core Complex : It continues the transcription.
(c) Rho Factor () : It terminates transcription, its molecular weight is 55,000.
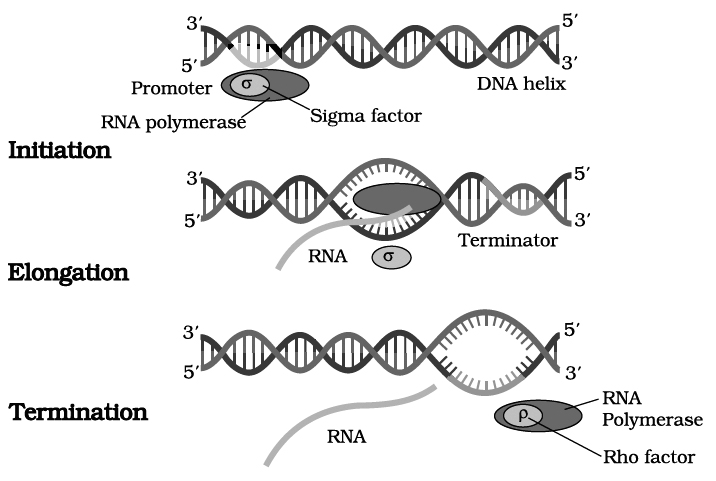
Transcription in Eukaryotes
In eukaryotes, the promotor site is recognised by presence of specific nucleotide sequence called TATA box or Hogness box (7 base pair long -TATATAT or TATAAAT) located 20 bp upstream to the start point.
Another sequence is CAAT box present between -70 and -80 bp.
In eukaryotes, the RNA polymerases are of three types, i.e., RNA polymerase I, RNA polymerase II and RNA polymerase III.
Functions of different RNA polymerases in eukaryotes are given below:
(i) RNA polymerase-I 5.8 S, 18 S, 28 S rRNA synthesis
(ii) RNA polymerase-II Hn RNA (heterogenous nuclear RNA), mRNA synthesis
(iii) RNA polymerase-III tRNA, Sc RNA (small cytoplasmic RNA), 5S rRNA and Sn RNA (small nuclear RNA) synthesis.
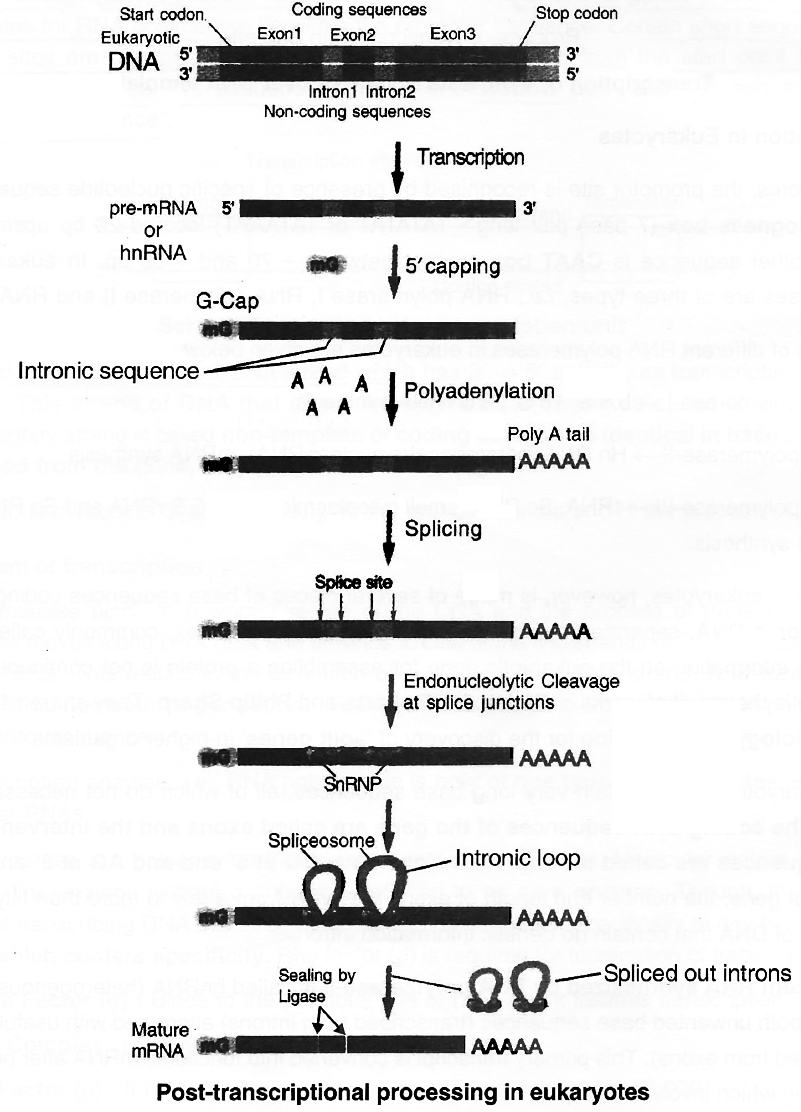
The gene in eukaryotes, however, is made of several pieces of base sequences coding for amino acids called exonic DNA, separated by stretches of non-coding sequences, commonly called intronic DNA.
Thus, the information on the eukaryotic gene for assembling a protein is not continuous but split.
This discovery is the result of works of Richard J. Roberts and Philip Sharp.
They shared 1993 Nobel Prize for Physiology and Medicine for the discovery of 'split genes' in higher organisms.
Most eukaryotic genes contain very long base sequences, all of which do not necessarily form mature mRNA.
The coding DNA sequences of the gene are called exons and the intervening non-coding DNA sequences are called introns.
All introns have GU at 5' end and AG at 3' end. Depending on the size of gene, the number and length of exons may vary from a few to more than fifty, alternating with stretches of DNA that contain no genetic information introns.
The nascent RNA synthesized by RNA polymerase-II is called hnRNA (heterogenous nuclear RNA).
It contains both unwanted base sequences (transcribed from introns) alternated with useful base sequences (transcribed from exons).
This primary transcript is converted into functional mRNA after post-transcriptional processing which involves 3 steps:
1. Modification of 5' end by Capping: Capping at 5' end occurs rapidly after the start of transcription.
The guanosine methylated at 7th position is added at 5' with the help of enzyme guanyl transferase.
Cap is essential for formation of mRNA-ribosome complex.
Translation is not possible if cap is lacking, because cap is identified by 18 S rRNA of ribosome unit.
2. Polyadenylation at 3' end (Tailing) : Poly (A) is added to 3' end of newly formed hn RNA with the help of enzyme Poly A polymerase. It adds about 200-300 adenyl ate residues.
3. Splicing of hnRNA (Tailoring) : In eukaryotes the coding sequences of RNA (exons) are interrupted by non coding sequences (introns).
Small nuclear RNA (snRNA) + Protein complex called small nuclear ribonucleo protein or SnRNPs (or snurps) play important role in this process.
Here the introns are removed and exons come in one plane.
This process is called splicing through which a mature mRNA is produced.
Normally, mRNA carries the codons of single complete protein molecule (monocistronic mRNA) in eukaryotes, but in prokaryotes, it carries codons from several adjacent DNA cistrons and becomes much longer in size (polycistronic mRNA).

 ACME SMART PUBLICATION
ACME SMART PUBLICATION
