- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
Human Genome Project (HGP)
The Human genome project was the international, collaborative research program whose goal was the complete mapping and understanding of all the genes of human beings.
All genes together are known as "genome".
The Human Genome Project as "Mega project" was a 13 year project co-ordinated by the U.S. Department of Energy and the National Institute of Health.
An International Human Genome project was launched in the year 1990 and completed in 2003.
The International Human Genome sequencing consortium published the first draft of the human genome in the journal Nature in February 2001 .
Human genome is said to have approximately 3 × 109 bp, and the cost of sequencing required is US $ 3 per bp. The total estimated cost of the project would be approximately 9 billion US dollars. In human genome 20,000 to 25,000 genes are present, out of them smallest gene is TDF gene with 14 bp and largest gene is Duchenne Muscular Dystrophy gene with 2400 × 103 bp.
Organisms and the number of bp and genes in them
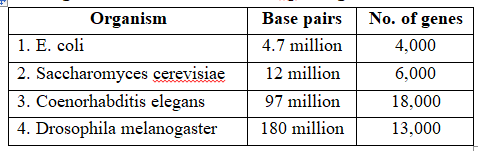
Following are the important goals of HGP :Goals of HGP
(i) Identication of all the approximately 20,000 -25,000 genes in human DNA.
(ii) To determine the sequence of the 3 billion chemical base pairs that make up human DNA.
(iii) To store this information in databases.
(iv) To improve tools for data analysis.
(v) Transfer related technologies to other sectors, such as industries.
(vi) Bioinformatics, i.e., close association of HGP with the rapid development of a new area in biology.
(vii) Sequencing of model organisms -Complete genome sequence of E.coli, Saccharomyces cerevisiae, Coenorhabditis elegans; Drosophila melanogaster, D. pseudoobscura, Oryza sativa and Arabidopsis was achieved in April, 2003.
(viii) Adress the ethical, legal and social issues (ELSI) that may arise from the project.
Concept Builder
Key Definitions :
1. cDNA: It stands for complementary DNA, a synthetic type of DNA generated from mRNA. By using mRNA as template, scientists use enzymatic reactions to convert its information back into cDNA and then clone it as cDNA library. These libraries are important to scientists because they consist of clones of all protein -encoding DNA or all the genes in the human genome.
2. Mb : Mb stands for megabase, a unit of length equal to 1 million base pairs and roughly equal to 1cM. (1cM = 1 million bp)
3. Microarray: Microarrays are devices used in many types of large scale genetic analysis. They can be used to study how large number of genes are expressed as messenger RNA in a particular tissue and how a cell's regulatory network control vast batteries of genes simultaneously.
Methodologies
The HGP techniques include:
(i) Sequence tagged site: It is a short DNA segment that occurs only once in a genome and whose exact location and order of bases is known. STSs serve as landmarks on the physical map of a genome. It is also called as Expressed Sequence Tags (ESTs). The genes that are expressed as RNA are referred to as ESTs.
(ii) Sequencing the whole set of genome that contained all the coding and non-coding sequences, and later assigning different regions in the sequence with functions, known as Sequence Annotation.
(iii) The Employment of Restriction Fragment Length Polymorphism (RFLP)
(iv) Yeast Artificial Chromosomes (YACs)
(v) Bacterial Artificial Chromosomes (BACs)
(vi) Polymerase chain reaction (PCR)
(vii) Electrophoresis
For sequencing, the total DNA from a cell is isolated and converted into random fragments of relatively smaller sizes (recall DNA is a very long polymer, and there are technical limitations in sequencin very long pieces DNA) and cloned in suitable host using specialised vectors.
The cloning resulted into amplification of each piece of DNA fragment so that it subsequently could be sequenced with ease.
The commonly used hosts were bacteria and yeast, and the vectors were called as BAC (bacterial artificial chromosomes), and YAC (yeast artificial chromosomes).
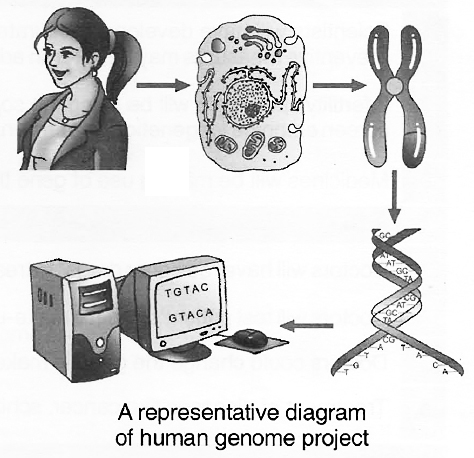
The fragments were sequenced using automated DNA sequencers that worked on the principle of a method developed by Frederick Sanger. These sequences were then arranged based on some overlapping regions present in them. This required generation of overlapping fragments for sequencing. Alignment of these sequences was humanly not possible. Therefore, specialised computer based programs were developed. These sequences were subsequently annotated and were assigned to each chromosome. The sequence of chromosome 1 was completed only in May 2006 (this was the last of the 24 human chromosomes -22 autosomes and X and Y -to be sequenced).
Salient Features of Human Genome
Some of the salient observations drawn from human genome project are as follows :
(i) The human genome contains 3164.7 million nucleotide bases.
(ii) The average gene consists of 3000 bases, but sizes vary greatly, with the largest known human gene being dystrophin at 2.4 million bases.
(iii) The total number of genes is estimated at 30,000-much lower than previous estimates of 80,000 to 1,40,000 genes. Almost all (99.9 percent) nucleotide bases are exactly the same in all people.
(iv) The functions are unknown for over 50 per cent of discovered genes.
(v) Less than 2 percent of the genome codes for proteins.
(vi) Repeated sequences make up very large portion of the human genome.
(vii) Repetitive sequences are stretches of DNA sequences that are repeated many times, sometimes hundred to thousand times. They are thought to have no direct coding functions, but they shed light on chromosome structure, dynamics and evolution.
(viii) Chromosome 1 has most genes (2968), and the Y has the fewest (231).
(ix) Scientists have identified about 1.4 million locations where single-base DNA differences (SNPs -single nucleotide polymorphism, pronounced as 'snips') occur in humans. This information promises to revolutionise the processes of finding chromosomal locations for disease-associated sequences and tracing numan history.
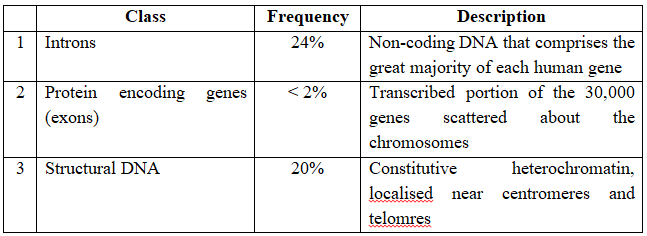
Classes of DNA sequences found in Human Genome
Francis Collins, director of the public funded genome project predicts the following progress in the next thirty years:Future Thrust of Human Genome Project
By 2010
1. Scientists will have developed accurate predictive tests for atleast a dozen common diseases so that preventive measures may be taken in advance.
2. Infertility specialists will be using the sophisticated techniques of pre-implantation genetic diagnosis to screen embryos for genetic disorders and designer babies.
3. Medicines will be making use of gene therapy.
By 2020
1. Doctors will have "designer drugs" to treat almost every disease.
2. Doctors will test patient's genetic make-up before prescribing drugs.
3. Doctors could change the genetic make-up of a living person by germ-line therapy.
4. Treatment of diseases like cancer, schizophrenia, depression etc. will have transformed.
By 2030
1. Genetic-based health care will have completely developed.
2. Most medical researches will be carried out using computer models rather than living tissues of animals.
3. Average life-span will rise up to 90 years.
The Indian Scenario
The Indian gene centre accounts for 160 species out of 2400 species in all of the 12 megagene centres.
Though the plant genetic resources activities got intensified in India in the first half of the century, but received the required impetus after creation of NBPGR in 1976 which has headquarters at IARI (Indian Agricultural Research Institute), New Delhi and several regional sub centres at different agroclimatic zones like Jodhpur (Arid areas), Trichur (Humid Tropical Zone), Shillong (Eastern sub tropical/subtemperate zone) etc.
As on 31 st March 1996, NBPGR holds 15,4,533 accessions of various agri-horticultural crops.
National Facility for Plant Tissue Culture Repository (NFPTCR) was established in 1986 by department of biotechnology at NBPGR for conservation of germplasm of vegetatively propagated plants.
Concept Builder
1. In translation, ribosomes are often found in clusters linked together by a strand of mRNA. This complex (polyribosome or polysome) enables several molecules of the same polypeptide to be produced simultaneously.
2. In vitro synthesis of gene was made by Hargobind Khorana (1968). He was able to link 77 nucleotide base pairs to code for yeast tRNAalanine. Later on in 1976, he synthesized a functional gene by linking 207 base pairs to code for E.coli. tRNA tyrosine.
3. Jumping genes (Transposon or Transposable element) discovered by Babara McClintock in maize can change their position within or between two chromosomes.
4. In some viruses a part of genetic material function for two or more genes (cistrons). Such genes are called overlapping genes. They were discovered by Borrel in × 174.
5. Transgenosis is the transmission of genetic information to plant cells by bacteriophage or phenomenon of transfer, gene maintenance, transcription and function.
6. Siblings (also sibship or sibs) : Individuals having the same parents; brother-sister relationship.
7. Genetic marker. A gene mutation that has phenotypic effects useful for tracing the chromosome on which it is located.
8. Phenocopy: An individual who has a phenotype similar to that produced by a certain mutant genotype, even though the individual may not have that genotype.
9. Charles Weismann isolated the gene (cDNA) for interferon from human lymphocytes and cloned it in E. coli and yeast.
10. Some Other Important Contributions of Scientists
(i) Zacharis - Called nuclein as DNA and proved that chromatin is rich in DNA
(ii) Fuelgen - DNA is located in nucleus where chromosomes are found, nucleus is fuelgen positive
(iii) Behrem - Nucleic acid is of two types DNA and RNA.
(iv) Fischer - Identified purines and pyrimidines in nucleic acid.
(v) Levene - Studied chemistry of DNA and found that DNA has deoxyribose sugar and four types of nucleotides.
(vi) Sanger - Determined base sequence in nucleic acid.
(vii) Hedges and Jacob -Term "transposon"

 ACME SMART PUBLICATION
ACME SMART PUBLICATION
