- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
REPLICATION OF DNA
The Watson-Crick model of DNA immediately suggested that the two strands of DNA would separate.
Each separated or parent strand serves as a template (model or guide) for the formation of a new but complementary strand.
Thus, the new or daughter DNA molecules formed would be made of one old or parental strand and another newly formed complementary strand.
This method of formation of new daughter DNA molecules is called semi-conservative method of replication.
The following experiment suggests that DNA replication is semi-conservative.
Messelson and Stahl (1958) conducted experiments using heavy nitrogen (15N) to determine whether the concept of semi-conservative replication is correct.
They used Cesium chloride (CsCl) gradient centrifugation technique for this purpose.
A dense solution of CsCl, on centrifugation, forms density gradient-bands of a solution of lower density at the top that increases gradually towards bottom with highest density.
If DNAs of different densities are mixed with CsCl solution, these would separate from one another and would form a definite density band in the gradient along with CsCl solution.
Messelson and Stahl created DNA molecules of different densities by using normal nitrogen 14N and its heavy isotope 15N.
For this purpose, Escherichia coli was grown in 15NH4Cl containing culture medium for many generations, so that bacterial DNA become completely heavy.
This non-radioactive or heavy DNA (incorporating 15N) had more density than DNA with normal nitrogen (14N).
The bacteria were then transferred to culture medium containing only normal nitrogen (14NH4Cl). The change in density was observed by taking DNA samples periodically.
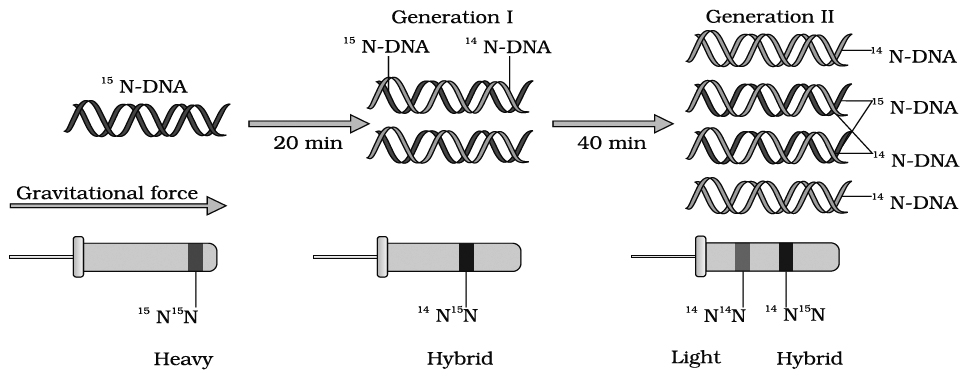
Messelson and Stahl's Experiment
If DNA replicates semi-conservatively, then each heavy (15N) DNA strands should separate and each separated strand should acquire a light (14N) partner after one round of replication.
This should be a hybrid DNA made of two strands, i.e., 14N–15N.
Meselson and Stahl observed that such DNA was actually half dense indicating the presence of hybrid DNA molecules.
After second round of replication there would be four DNA molecules.
Of these, two molecules would be hybrid (14N–15N) showing half density as earlier and the remaining two molecules would be made of light strands (14N–14N) Thus, after second generation, the same half dense band (14N–15N) was seen but the density of light bands (14N–14N) increased.
Messelson and Stahl's work as such provided confirmation of Watson-Crick model of DNA and its semi-conservative replication.
Taylor proved semiconservative mode of chromosome replication in eukaryotes using tritiated thymidine in the root of Vicia faba (faba beans).
Cairns proved semiconservative mode of replication in E. coli by using tritiated thymidine (H3–tdR) in autoradiography experiment.
He proposed -model for replication in circular DNA.
Mechanism of DNA Replication
DNA replication involves following four major steps:
(1) Initiation of DNA replication
(2) Unwinding of helix
(3) Formation of primer strand
(4) Elongation of new strand
1. Initiation of replication.
Replication of DNA always begins at a definite site called origin of replication.
Prokaryotes have single origin of replication.
It is called ori-c in E.coli.
On the other hand, eukaryotes have several thousand origins of replication.
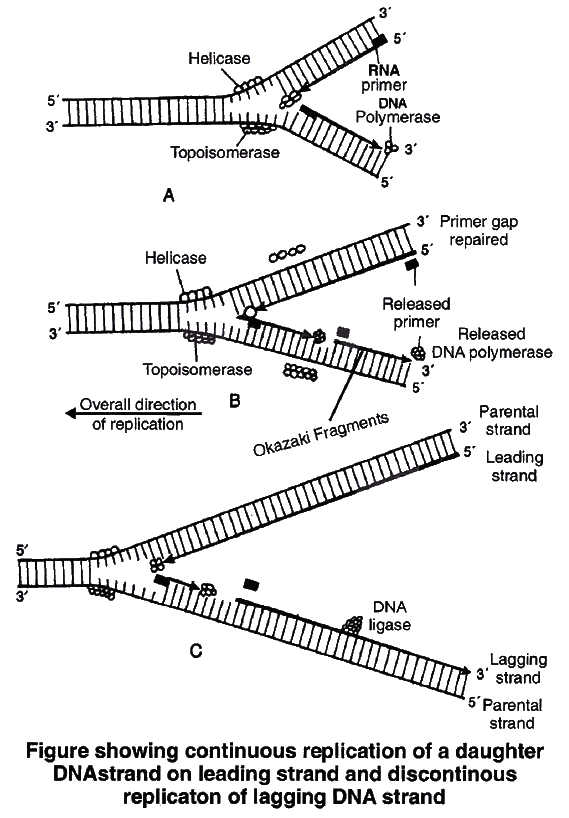
2. Unwinding of helix.
DNA replication requires that a double helical parental molecule is unwound so that its internal bases are available to the replication enzymes.
Unwinding is brought about by enzyme helicase which is ATP dependent.
Unwinding of DNA molecule into two strands results in the formation of Y-shaped structure, called replication fork.
These exposed single strands are stabilised by a protein known as Single-strand binding protein (SSB).
Due to unwinding, a supercoiling develops on the end of DNA opposite to replicating fork.
This tension is released by enzyme topoisomerase.
3. Formation of primer strand.
A new strand is to be synthesized opposite to the parental strands DNA polymerase III is the true replicase in E. coli, which is incapable of initiating DNA synthesis, i.e., it is unable to deposit the first nucleotide in a daughter (new) strand without the Primer.
Another enzyme, known as primase, strand synthesizes a short primer strand of RNA.
The primer strand then serves as a stepping stone (to start errorless replication).
Once the initiation of DNA synthesis is completed, this primer RNA strand is then removed enzymatically.
4. Elongation of new strand.
Once the primer strand is formed, DNA replication occurs in 53 direction, i.e., during synthesis of a new strand, deoxyribonucleoside triphosphates (dATP, dGTP, dTTP, dCTP) are added only to the free 3OH end.
Thus, the nucleotide at 3 end is always the most recently added nucleotide to the chain.
As the DNA replication proceeds on the two parental strands, synthesis of daughter or new strand occurs continuously along the parent 3'5' strand.
It is now known as leading daughter strand.
Synthesis of another daughter strand along the other parental strand, however, takes place in the form of short pieces.
This is called lagging daughter strand. These short pieces of DNA are known as Okazaki fragments, these segments are about 1,000-2,000 nucleotides long in prokaryotes.
Hense, DNA replication is semidiscontinuous. Discontinuous pieces of the lagging strand are joined together by the enzyme DNA ligase (after removal of primer) to form continuous daughter strand.
Thus two DNA molecules are now formed from one molecule.
Each of these daughter DNA molecules is made of two strands, of which one is old (parental) and other one is new or complementary strand.
DNA polymerase is the most important enzyme of DNA replication.
DNA polymerases are of three types i.e. DNA polymerase I, II and III in prokaryotes.

DNA polymerase II & III have only 3' 5' exonuclease activity.
Kornberg (1956), succeeded in demonstrating the in vitro synthesis of DNA molecule using a single strand of DNA as a template.
He extracted and purified an enzyme from E. coli which was capable of linking free DNA nucleotides, in presence of ATP as an energy source, to form complementary strand.
He called it DNA polymerase. DNA polymerase-I is called Kornberg enzyme.
In eukaryotes DNA polymerases are of 5 type these are DNA polymerase , , , and .
In eukaryotes, the replication of DNA takes place at s-phase of the cell cycle. The replication of DNA and cell division cycle should be highly co-ordinated. A failure in cell division after DNA replication result into polyploidy (achromosomal anomaly).
STRUCTURE OF RNA (Ribose Nucleic Acid)
RNA or ribonucleic acid is present in all the living cells. It is laevo rotatory and is responsible for learning and memory. It is found in the cytoplasm as well as nucleus. Description of types and structure of RNA is given below.
Types of RNA
In bacteria, there are three major types of RNAs: mRNA (messenger RNA), tRNA (transfer RNA), and rRNA (ribosomal RNA).
All three RNAs are needed to synthesise a protein in a cell.
The mRNA provides the template, tRNA brings aminoacids and reads the genetic code, and rRNAs play structural and catalytic role during translation.
RNA is generally involve in protein synthesis but in majority of plant viruses, it serves as a genetic material. Therefore, there are two major types of RNA-(A) genetic RNA and (B) non-genetic RNA.
(A) Genetic RNA -
Fraenkel-Conrat showed that RNA present in TMV (Tobacco Mosaic Virus) is a genetic material. Since then, it is established that RNA acts as a genetic material in most plant viruses.
(B) Non-genetic RNA.
This type of RNA is commonly present in cells where DNA is genetic material. Non-genetic RNA is synthesized on DNA template. It is of following three major types:
(a) Messenger RNA (mRNA).
It was reported by Jacob and Monad.
It carries genetic information present in DNA, so is called working copy.
mRNA constitutes about 3-5% of the total RNA present in the cell.
The molecular weight varies from 25,000 to 1,00,000.
It is about 300 nucleotide long at minimum.
Structure of prokaryotic m-RNA. It is polycistronic in nature i.e., several cistrons (functional part of DNA) form a single m-RNA. Thus, each m-RNA has a message to produce several polypeptides. Average life is 2 minutes.
At 5' end near initiation codon a sequence of fixed bases is found called Shine Dalgarno(SD) sequence (5'AGGAGGU3'). It helps in correct binding of ribosomal subunit (30 S) on it.
Structure of eukaryotic m-RNA. It is monocistronic in nature and has a message to produce on polypeptide only. They are metabolically stable and their life varies from few hours to days.

UTR (Untranslated region). They are sequences of RNA before start or initiation codon and after stop or termination codon. They are not translated. They are transcribed as part of same transcript as the coding region. Such UTRs provide stability to m-RNA and also increase translational efficiency.
(b) Ribosomal RNA (rRNA). It was reported by Kuntz. It is most stable type of RNA and is a constituent of ribosomes. It forms about 80% of the total cellular RNA.
In eukaryotes, 4 types of rRNA's found are 28 S, 18 S, 5.8 S and 5 S whereas, in prokaryotes three types of rRNA's found are 23 S, 16 S and 5 S. It is synthesised by genes present on DNA of several chromosomes found within a region known as nucleolar organiser.
(c) Transfer RNA (tRNA). The existence of tRNA was postulated by Crick. It is also known as soluble RNA (sRNA). These are the smallest molecules which carry amino acids to the site of protein synthesis. These constitute about 15% of the total cellular RNA.
It acts as an intermediate molecule between triplet code of mRNA and amino acid sequence of polypeptide chain. All tRNA's have almost same basic structure. There are over 60 types of tRNA.
Structure of t-RNA
Clover leaf model of tRNA is a 2-dimensional model suggested by Holley et.al. tRNA molecule appears like a clover leaf, being folded with three or more double helical regions (stem), having loops also. The three dimensional structure of the tRNA was proposed to be inverted L-shaped by Kim and Klug.
All tRNA molecules commonly have a guanine residue at its 5' terminal end. At its 3' end, unpaired – CCA sequence is present. Amino acid gets attached at this end only. The number of nucleotide varies from 77 (tRNAalanine) to 207 (tRNAtyrosine).
There are three loops in t-RNA
(i) Amino acyl synthetase binding loop, also called DHU loop.
(ii) Ribosomal binding loop with 7 unpaired bases. It is also called TC loop.
(iii) Anticodon loop with 7 unpaired bases. Out of the 7 bases in anticodon loop, 3 bases acts as anticodon for a particular triplet codon present on mRNA.
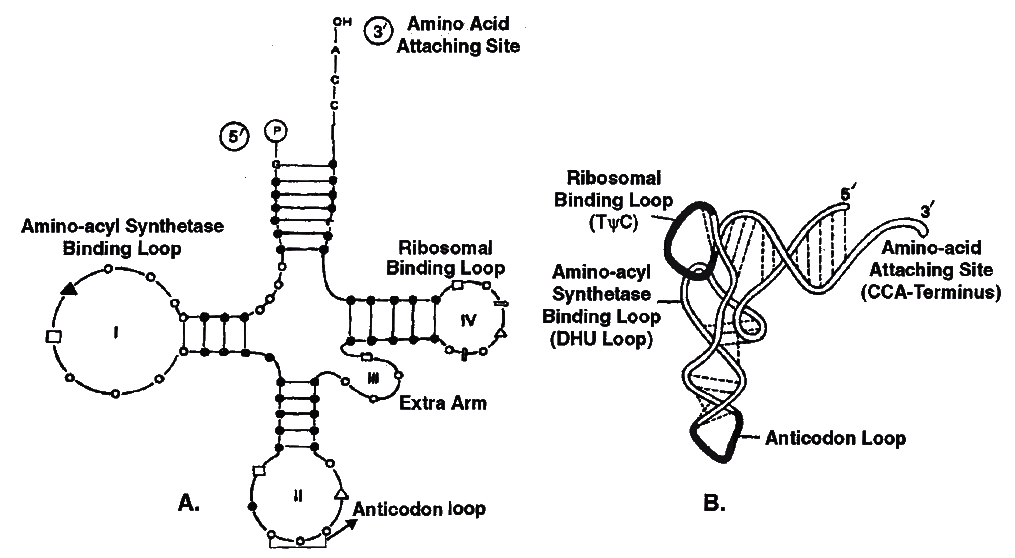
Structure of tRNA
A. Clover leaf model to show basic plan of tRNA secondary structure or 2D structure
B. Three Dimensional Structure showing L-shaped configuration
GENE EXPRESSION
DNA, being the genetic material, carries all the informations necessary to programme the functions of a cell by controlling the synthesis of enzymes or proteins.
Beadle and Tatum put forward a theory-one gene one enzyme in support of the earlier hypothesis that enzymes are proteinaceous in nature and each is produced by a single gene.
They conducted experiments on the nutritional strains of pink mould, Neurospora crassa.
This fungus grows on simple nutrient medium and has the ability to synthesize all its cellular components.
Such an organism is called prototroph.
An organism that is unable to synthesize a particular cellular metabolite, such as an amino acid or coenzyme is called auxotroph.
Beadle and Tatum produced arginine (an amino acid) auxotrophs (mutants of Neurospora unable to synthesize arginine) by giving X-rays treatment to the cells. Arginine synthesis passes through the following path

They found that any step of this metabolic chain could be blocked by a mutation in a specific enzyme catalyzing the reaction, each enzyme representing a different gene product.
Thus, Beadle and Tatum reached a conclusion that each gene functions to produce a single enzyme.
Some proteins, e.g., haemoglobin and other quaternary proteins are made up of two or more than two polypeptide chains. After it, the 'one gene one enzyme' theory was modified into one gene one polypeptide hypothesis by Yanofsky. Later Jacobson and Baltimore proposed one mRNA -one polypeptide hypothesis.
Gene and protein : A gene expresses itself by protein synthesis.
When a particular gene is expressed (i.e., controls a function or a reaction), its information is copied into another nucleic acid, i.e., mRNA (messenger RNA) which in turn directs the synthesis of specific proteins.
These concepts form the central dogma of molecular biology. This has been shown by F. Crick in the following diagram.
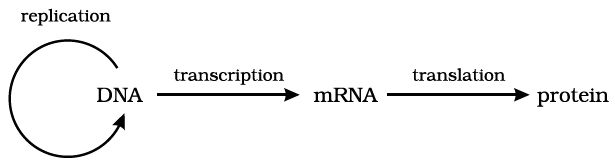
An exception to this one way flow of information was reported in 1970.
H. Temin and D. Baltimore independently discovered reverse transcription in some viruses.
These viruses can code an enzyme reverse transcriptase which can code DNA on RNA template.
This discovery was important in understanding cancer and, hence, these two scientists were awarded Nobel Prize. The modified flow of information now can be shown as below:

 Reverse flow of transcriptional information or Reverse Central Dogma or Teminism
Reverse flow of transcriptional information or Reverse Central Dogma or Teminism
Commoner suggests circular flow of information.

 ACME SMART PUBLICATION
ACME SMART PUBLICATION
