- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
REGULATION OF GENE EXPRESSION
1. Constitutive genes are those genes which are constantly expressing themselves in a cell because their products are required for the normal cellular activities, e.g., genes for glycolysis, ATPase.
2. Non-constitutive genes are not always expressing themselves in a cell. These are called Luxury genes. They are switched on or off according to the requirement of cellular activities, e.g., gene for nitrate reductase in plants, lactose system in Escherichia coli.
This provides maximum functional efficiency to the cell. Such regulated genes, therefore are required to be switched 'on' and 'off' when a particular function is to begin or stop.
This regulation can be achieved by anyone of the following two processes:
(1) Induction. This is a process of gene regulation where addition of a substrate stimulates or induces synthesis of enzymes needed for its own breakdown.
(2) Repression. In this process of gene regulation, addition of end product stops the synthesis of enzymes needed for its formation. This phenomenon is also known as feedback repression.
Operon
Francois Jacob and Jacques Monod (1961) proposed a model of gene regulation, known as Operon model.
Operon is a co-ordinated group of genes such as structural genes, operator gene, promoter gene, regulator gene which function or transcribe together and regulate a metabolic pathway as a unit.
Inducible operon (lac operon)
In Escherichia coli, breakdown of lactose requires three enzymes.
These enzymes are synthesized together in a co-ordinated manner and the unit is known as lac operon.
Since the addition of lactose itself stimulates the production of required enzymes, it is also known as inducible system.
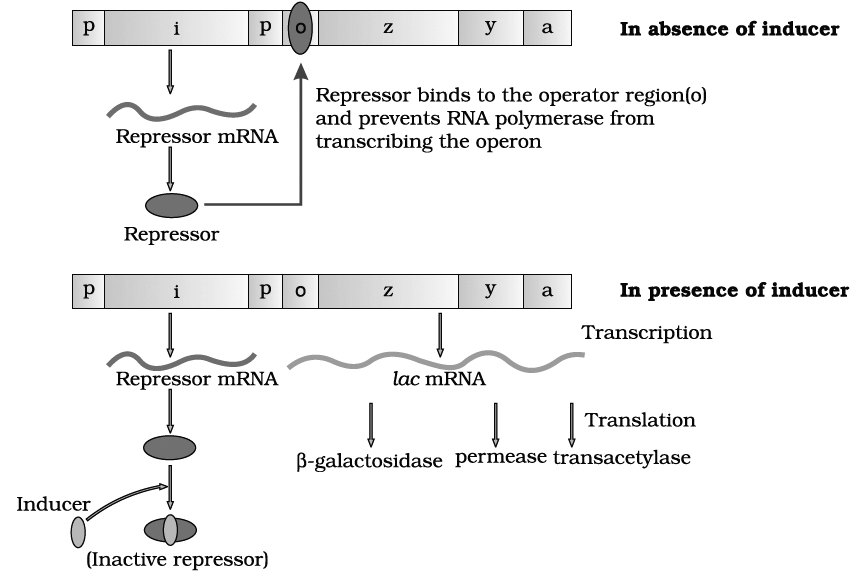
Lac operon
lac operon consists of following genes :
(1) Structural genes. These genes code for the proteins needed by the cell which include enzymes or other proteins having structural functions.
In lac-operon, there are following three structural genes:
(i) lac a -gene coding for enzyme transacetylase
(ii) lac y -gene coding for enzyme permease
(iii) lac z -gene coding for enzyme -galactosidase
(2) Operator gene (o). It interacts with a protein molecule (the regulator molecule), which promotes (induce) or prevents (repress) the transcription of structural genes.
(3) Promoter gene (p). This gene is the recognition point where RNA polymerase remains associated.
(4) Regulator gene (i). This is generally known as inhibitory gene (i). This gene codes for a protein called the repressor protein. It is an allosteric protein which can exist in two forms. In one form, it is paratactic to an operator and binds with it and in another form it is paratactic to inducer (like lactose).
The operon is switched 'off' and 'on'.
The transcription or function of lac genes or structural genes depends upon operator gene.
When repressor protein produced by inhibitory gene (i) or regulatory gene binds to operator (o) gene, RNA polymerase gets blocked. There would be no transcription and the operon model remains in 'switched off' position.
On the other hand, when inducer like lactose is added, the repressor protein (produced by gene i) gets bound to it.
The repressor, therefore, gets released from the operator.
RNA polymerase is now permitted to act.
Hence, the transcription of lac genes now takes place and the operon model is in 'switched on' position.
All the three genes are transcribed by RNA polymerase to form a single mRNA strand.
Each gene segment of mRNA is called cistron and, therefore, mRNA strand transcribed by more than one gene is known as polycistronic.
Tryptophan Operon -Repressible Operon System
Operon model can also be explained using feed-back repression.
In tryptophan (trp) operon, three enzymes are necessary for the synthesis of amino acid tryptophan.
These enzymes are synthesized by the activity of five different genes in a co-ordinated manner.
The addition of tryptophan, however, stops the production of these enzymes.
Thus, the system is known as repressible system.
In this system, there are five structural genes, trp A, trp B, trp C, trp D and trp E, coding for three enzyme needed for the synthesis of tryptophan, an amino acid.
Regulatory gene (R) produces repressor protein which is known as apo-repressor because it does not get bound to the operator directly.
Hence, the operator gene remains in 'switched on' position.
Tryptophan when added, binds to the apo-repressor and is called co-repressor.
This apo-repressor and co-repressor complex (activated repressor) now binds to operator gene and blocks the function of RNA polymerase.
Thus, transcription would not occur and tryptophan operon would be in 'switched off' position.
Feed-back repression is functional when there is no further need of the end product and, hence, there is no requirement of continuation of this anabolic pathway.
This operon stops the process.
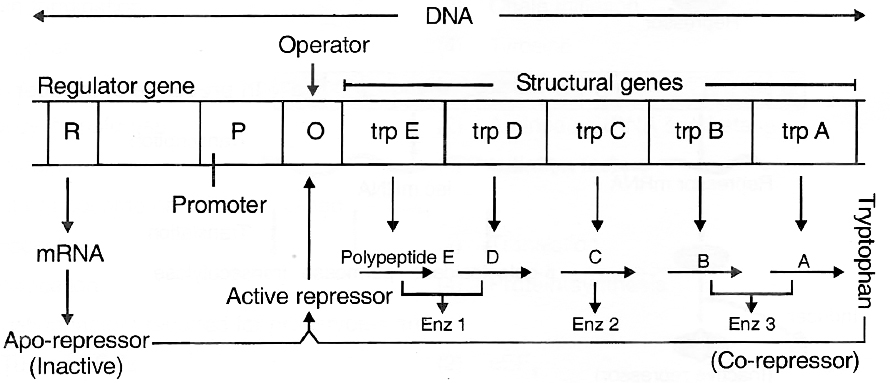
Tryptophan operon model of gene regulation in bacteria
In the prokaryotes, control of the rate of transcriptional initiation is the predominant site for control of gene expression.
Regulation of Gene Expression in Eukaryotes
In eukaryotes, the regulation of gene expression could be exerted at four levels:
(i) Transcriptional level (formation of primary transcript)
(ii) Processing level (regulation of splicing)
(iii) Transport of mRNA from nucleus to the cytoplasm
(iv) Translational level
In eukaryotes, functionally related genes do not represent an operon but are present on different sites in chromosomes.
Here, structural gene is called split gene which is a mosaic of exons and introns, i.e., base triplet -amino acid matching is not continuous.
Britten -Davidson gene battery model is most popular for eukaryotic genes.
It proposes the occurrence of 5-types of genes-producer, receptor, integrator, sensor and enhancer-silencer.
Exons are coding part of cistron that forms RNA.
Introns are non-translated part of DNA called IVS (intervening sequences) or spacer DNA.
An intron begins with GU and ends up with an AG.
However, the entire split gene is transcribed to form a continuous strip of hn-RNA.
This removal of non-coding intronic part and fusion of exonic coding parts of RNA is called RNA splicing. About 50-90% of primary transcribed RNA is discarded during processing.

 ACME SMART PUBLICATION
ACME SMART PUBLICATION
