- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
DNA FINGERPRINTING
What is DNA Fingerprinting?
The chemical structure of everyone's DNA is the same.
The only difference between people (or any animal) is the order of the base pairs.
There are so many millions of base pairs in each person's DNA that every person has a different sequence.
Using these sequences, every person could be identified solely by the sequence of their base pairs.
However, there are so many millions of base pairs due to which the task would be very time consuming.
Instead, scientists are able to use a shorter method, because of repeating base patterns in DNA (Satellite DNA).
These patterns do not, however, give an individual "fingerprint," but they are able to determine whether two DNA samples are from the same person, related people, or non-related individuals.
VNTR's, RFLP, SSR, RAPD
Every strand of DNA has stretches that contain genetic information which are responsible for an organism's development (exons) and stretches that, apparently, supply no relevant genetic information at all (introns).
Although the introns may seem useless, it has been found that they contain repeated sequences of base pairs.
These sequences are called Variable Number Tandem Repeats (VNTRs).
VNTR's also called minisatellites were discovered by Alec Jeffreys et. al. of U.K.
These consist of hypervariable repeat regions of DNA having a basic repeat sequence of 11-60 bp and flanked on both sides by restriction sites.
The number of repeats show a very high degree of polymophism and the size of VNTR varies from 0.1 to 20 Kb. The length of minisatellites and position of restriction sites is different for each person.
Therefore, when the genomes of two people are cut using the same restriction enzyme, the length and number of fragments obtained is different for both.
This is called RFLP or Restriction Fragment Length Polymorphism.
These fragments, when separated by gel electrophoresis, and obtained on a Southern Blot, constitutes what is called DNA fingerprint.
Father of DNA fingerprinting is Alec Jeffreys while the Indian experts Lalji Singh and V.K. Kashyap are known as Father of Indian technique.
Concept Builder
Now-a-days RAPD's or Randomly Amplified Polymorphic DNA are used for DNA fingerprinting (pronounced 'rapid').
It is a simpler technique than RFLP.
The DNA sequences flanking microsatellites (or SSRs i.e., simple sequence repeats or short tandem repeats) on both sides are conserved, so one can select primers complementary for these sequences and put these along with the genome in a thermal cycler.
The PCR will amplify the intervening microsatellites which can also be radiolabelled by using 32p.
The microsatellites have simple sequences of 1-6 bp. repeated hundreds of times.
Methodology of DNA fingerprinting
The Southern Blot is one way to analyze the genetic patterns which appear in a person's DNA. It was devised by E.M.Southern (1975) for separating DNA fragments. Performing a Southern Blot involves:
1. Isolating the desired DNA. It can be done either chemically, by using a detergent to wash the extra material from the DNA, or mechanically, by applying a large amount of pressure in order to "squeeze out" the DNA.
2. Cutting the DNA into several pieces of different size. This is done using one or more restriction enzymes.
3. Sorting the DNA pieces by size. The process by which the size separation is done is called gel electrophoresis. The DNA is poured into a gel, such as agarose, and an electrical charge is applied to the gel, with the positive charge at the bottom and the negative charge at the top. Because DNA has a slightly negative charge, the pieces of DNA will be attracted towards the bottom of the gel. The smaller pieces, however, will be able to move more quickly and thus, further towards the bottom than the larger pieces. The different-sized pieces of DNA will therefore, be separated by size, with the smaller pieces-towards the bottom and the larger pieces towards the top.
4. Denaturing the DNA fragments, so that all of the DNA is rendered single-stranded. This can be done either by heating or chemically treating the DNA in the gel using alkali.
5. Blotting the DNA. The gel with the size-fractionated. DNA is applied to a sheet of nitrocellulose paper or nylon membrane, and then baked to permanently attach the DNA to the sheet. The Southern Blot is now ready to be analyzed.
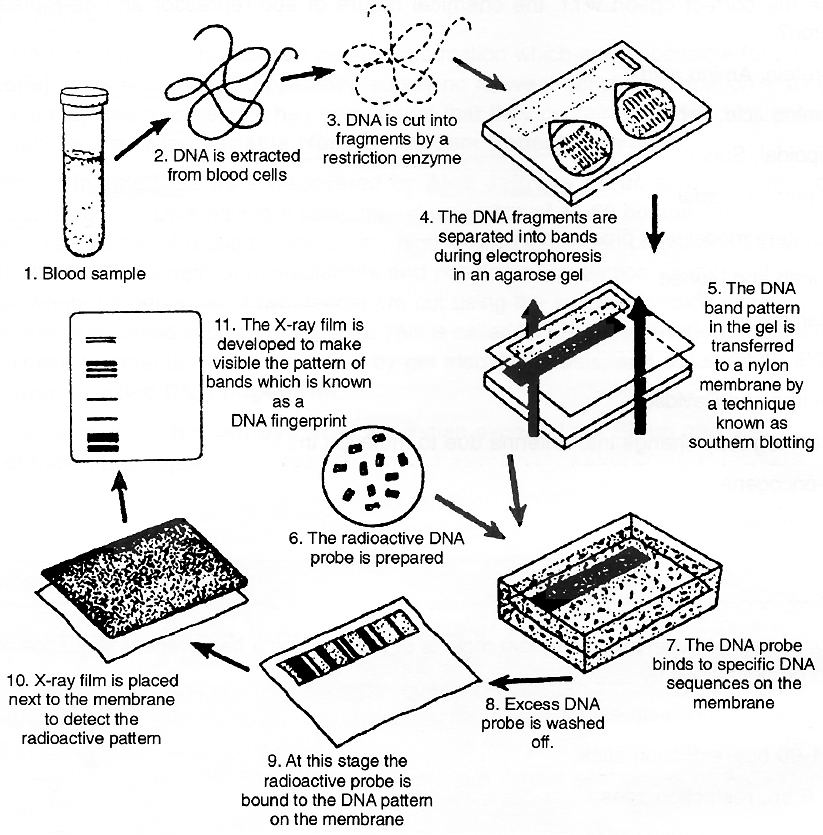
The DNA fingerprinting process
In order to analyze a Southern Blot, a radioactive genetic probe is used in a hybridization reaction with the DNA in question. If an X-ray is taken of the Southern Blot after a radioactive probe has been allowed to bound with the denatured DNA on the paper, only the areas where the radioactive probe binds will Show themselves on the film (autoradiography). This allows researchers to identify, in a particular person's DNA, the occurrence and frequency of the particular genetic pattern contained in the probe.
Practical Applications of DNA Fingerprinting
1. Solving Cases of Disputed Paternity and Maternity: Because a person inherits his or her VNTRs from his or her parents, VNTR patterns can be used to establish paternity and maternity.
2. Criminal Identification and Forensics: DNA isolated from blood, hair, skin cells, or other genetic evidence left at the scene of a crime can be compared, through VNTR patterns, with the DNA of a criminal suspect to determine guilt or innocence.
3. Personal Identification : The notion of using DNA fingerprints as a sort of genetic bar code to identify individuals has been discussed, but this is not likely to happen anytime in the near future. The technology required to isolate, keep on file, and then analyze millions of very specified VNTR patterns is both expensive and impractical.
GENOMICS
Application of DNA sequencing and genome mapping to the study, design and manufacture of biologically important molecules is known as genomics.
It is comparatively more recent branch in the field of biology.
The term genomics was introduced by Thomas Roderick.
In genomics, function of gene is identified by using the technique of reverse genetics.
Genomics study may be classified into three classes:
(i) Structural genomics : It involves mapping and sequencing of genes.
(ii) Functional genomics : It involves the identification of function of a particular gene.
(iii) Application genomics : It involves use of genomics information for crop improvement etc.
Complete genomics of Arabidopsis and rice have been worked out and they are having 130 and 430 million base pairs respectively.

 ACME SMART PUBLICATION
ACME SMART PUBLICATION
