- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
Translation
It is the mechanism by which the triplet base sequences of m-RNA molecules are converted into a specific sequence of amino acids in a polypeptide-chain. It occurs on ribosomes.
The major steps are:
(a) Activation of amino acids. In the presence of enzyme aminoacyl -tRNA synthetase (E), specific amino acids (AA) bind with ATP
![]()
 (b) Charging of t-RNA. The AA1-AMP-E1 complex formed in first step reacts with a specific tRNA. Thus, amino acid is transferred to tRNA. As a result, the enzyme and AMP are liberate
(b) Charging of t-RNA. The AA1-AMP-E1 complex formed in first step reacts with a specific tRNA. Thus, amino acid is transferred to tRNA. As a result, the enzyme and AMP are liberate
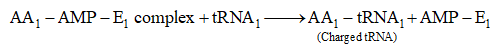
(c) Formation of polypeptide chain. It is completed in three steps.
(i) Chain Initiation. It requires 3 initiation factors in prokaryotes i.e., IF3, IF2, IF1.
9 initiation factors are required in eukaryotes i.e., eIF2, eIF3, eIF1, eIF4A, eIF4B, eIF4C, eIF4D, eIF5, eIF6.
(a) Binding of mRNA with smaller subunit of ribosomes (30S/40S). IF3 is involved in prokaryotes, while eIF2 is involved in eukaryotes. It involves interaction between Shine Delgarno sequence and 3' end of 16 S rRNA in prokaryotes.
(b) Binding of 30S / 40S-mRNA complex with t-RNA. Non-formylated methionine is attached with tRNA in eukaryotes and formylated methionine in prokaryotes. IF2 is involved in prokaryotes and eIF3 in eukaryotes.
![]()
(c) Attachment of larger subunit of ribosomes.
Initiation factors in eukaryotes -eIFl, eIF4.
Initiation factors in prokaryotes -IF1.
(ii) Chain elongation.
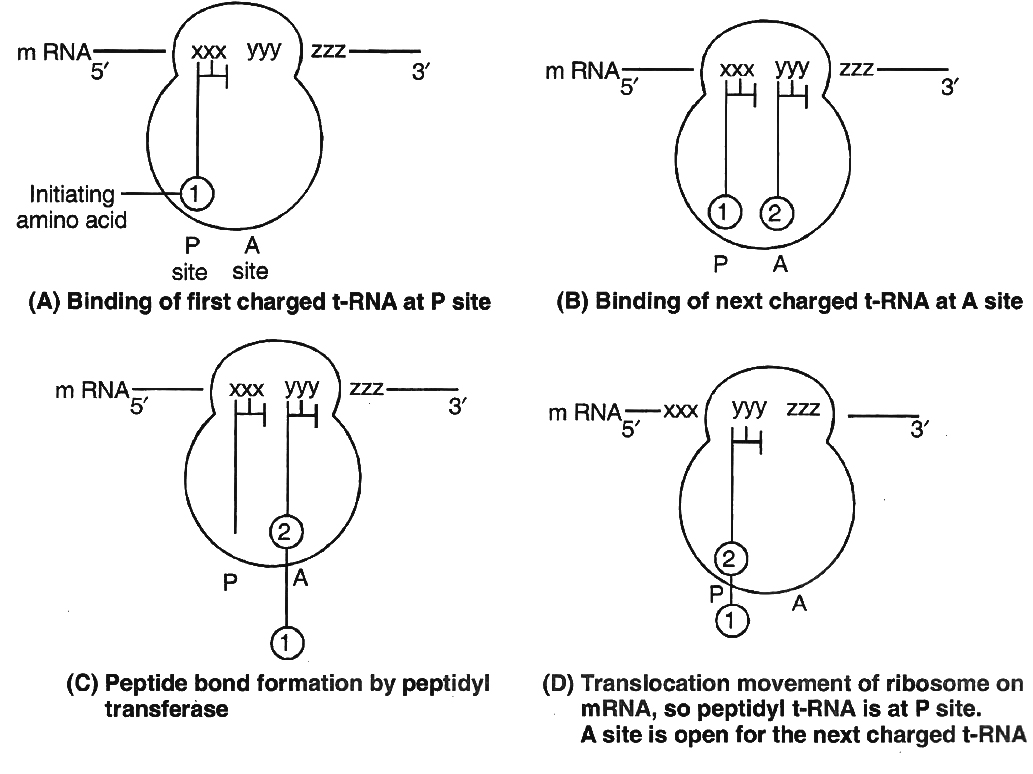
Ribosomes have two sites for binding amino acyl tRNA (i) Amino acyl or A site (acceptor site) (ii) Peptidyl site or P site (donor site).
A second charged tRNA molecule along with its appropriate amino acid approaches the ribosome at A site close to the P site.
Its anticodon binds to complementary codon of mRNA chain.
A peptide bond is formed between COOH group of first amino acid (methionine) and NH2 group of second amino acid.
The formation of peptide bond requires energy and is catalysed by enzyme peptidyl transferase. The initiating formyl methionine or methionine tRNA can bind only with P site.
All other newly coming aminoacyl tRNA bind to A site.
The elongation factor required for prokaryotes are EF-Tu and EF-Ts and in eukaryotes eEF1.
Translocation is movement of ribosome on mRNA. It requires EF -G in prokaryotes and eEF2 in eukaryotes.
(iii) Chain termination. The termination of polypeptide is signalled by one of the three terminal codons in the mRNA. The three terminal codons are UAG (Amber), UAA (Ochre) and UGA (Opal).
A GTP dependent factor known as release factor is associated with termination codon. It is eRF1 in eukaryotes and RF1 and RF2 in prokaryotes.
At the time of termination, the terminal codon immediately follows the last amino acid codon. After this, the polypeptide chain, tRNA, mRNA are released and the subunits of ribosomes get dissociated.
Concept Builder
There are some metabolic inhibitors which inhibit the synthesis of proteins in bacteria. These antibiotics are used in checking the growth of certain bacteria which are pathogenic in nature e.g.,
Tetracycline - Inhibits binding of amino-acyl tRNA to ribosomes
Streptomycin - Inhibits initiation of translation and causes misreading
Choloramphenicol - Inhibits peptidyl transferase activity
Erythromycin - Inhibits translocation of mRNA along ribosomes
Neomycin - Inhibits interaction between tRNA and mRNA.
Gene expression is the mechanism at the molecular level by which a gene is able to express itself in the phenotype of an organism.
In the translation unit, mRNA has some sequences that are not translated and are referred to as untranslated regions (UTR).
The UTRs are present at both 5' end (before start codon) and at 3' end (after stop codon).
They are required for efficient translation process.

 ACME SMART PUBLICATION
ACME SMART PUBLICATION
