- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
The DNA
DNA is a long polymer of deoxyribonucleotides.
The length of DNA is usually defined as number of nucleotides or a pair of nucelotide referred to as base pairs present in it.
This also is the characteristic of an organism.
Few examples are sited below :

Chemical structure of DNA polynucleotide :
The basic unit of DNA is a nucleotide which has three components -a nitrogenous base, a pentose sugar (deoxyribose), and a phosphate group.Chemical structure of DNA polynucleotide :
There are two types of nitrogenous bases - Purines (Adenine and Guanine), and Pyrimidines (Cytosine and Thymine).
Cytosine is common for both DNA and RNA and Thymine is present in DNA.
Uracil is present in RNA at the place of Thymine.
A nitrogenous base is linked to the pentose sugar through a N-glycosidic linkage to form a nucleoside, such as adenosine or deoxyadenosine, guanosine or deoxyguanosine, cytidine or deoxycytidine and deoxythymidine.
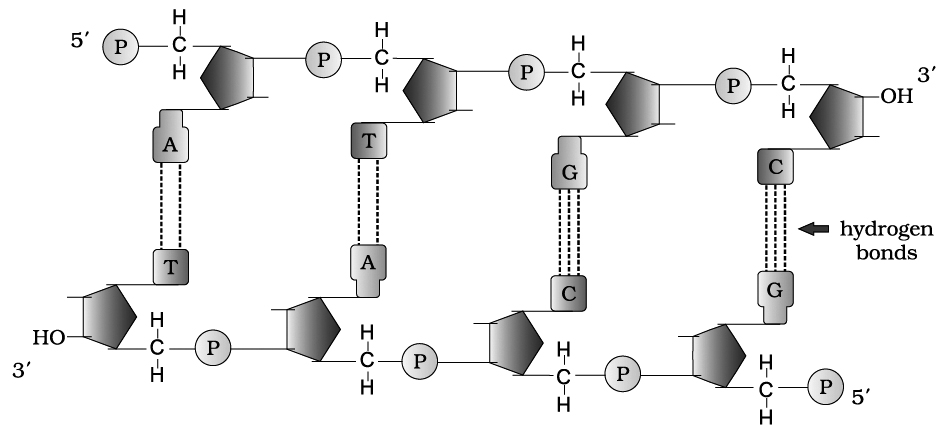
When a phosphate group is linked to 5'-OH of a nucleoside through phosphoester linkage, a corresponding nucleotide (or deoxynucleotide depending upon the type of sugar present) is formed.
Two nucleotides are linked through 3'-5' phosphodiester linkage to form a dinucleotide.
More nucleotides can be joined in such a manner to form a polynucleotide chain.
A polymer thus formed, has a free phosphate moiety at 5'-end of sugar, which is referred to as 5'-end of polynucleotide chain.
Similarly, at the other end of the polymer the sugar has a free 3'-OH group which is referred to as 3'-end of the polynucleotide chain.
The backbone in a polynucleotide chain is formed due to sugar and phosphates (phosphodiester bond) nitrogenous bases linked to sugar moiet projects from the backbone.
In RNA, every nucleotide residue has an additional -OH group present at 2' -position in the ribose.
Also, in RNA the uracil is found at the place of thymine (S-methyl uracil, another chemical name for thymine).
In 1953, James Watson and Francis Crick based on the X-ray diffraction data produced by Maurice Wilkins and Rosalind Franklin, proposed Double Helix model for the structure of DNA.
One of the hallmarks of their proposition was base pairing between the two strands of polynucleotide chain.
However, this proposition was also based on the observation of Erwin Chargaff –that for a double stranded DNA, the ratios between Adenine and Thymine and Guanine and Cytosine are constant and equal.
Chargaff (1950) made observations on the bases and other contents of DNA. These observations or generalizations are called Chargaff's rules.
(i) Purine and pyrimidine base pairs are in equal amount, i.e., adenine + guanine = thymine + cytosine.
(ii) Molar amount of purine (adenine) is always equal to the molar amount of pyrimidine (thymine). Similarly, guanine is equalled by cytosine.
(iii) Sugar deoxyribose and phosphate occur in equimolar proportions.
(iv) The ratio of A + T/G + C is constant for a species (Base ratio, e.g., 1.52 for human and 0.93 for E.coli.
The base pairing is a very unique property of the polynucleotide chains.
They are said to be complementary to each other, and therefore if the sequence of bases in one strand is known then the sequence in other strand can be predicted.
Thus, if one DNA strand has A, the other would have T and if one has G, the other, would have C.
Therefore, if the base sequence of one strand is CAT TAG GAC, the base sequence of other strand would be GTA ATC CTG.
Hence, the two polynucleotide strands are called complementary to one another.
Also, if each strand from a DNA or parental DNA acts as a template for synthesis of a new strand, the two double stranded DNA or daughter DNA produced would be identical to the parental DNA molecule.
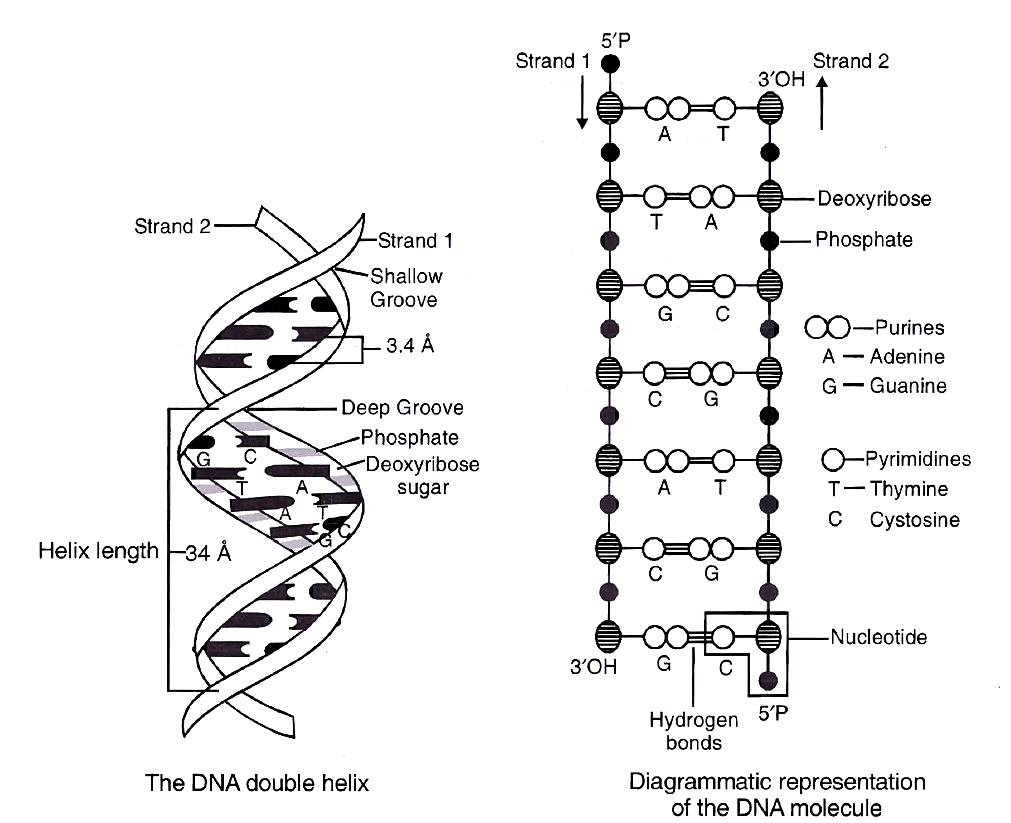
Salient features of DNA Double Helix
(i) It is made of two polynucleotide chains, where the backbone is constituted by sugar-phosphate, and the bases project inside.
(ii) The two chains have anti-parallel polarity. It means, if one chain has the polarity 5P 3OH, the other has 3OH 5P.
(iii) The bases in two strands are paired through hydrogen bond (H-bonds), forming base pairs(bp). Adenine forms two hydrogen bonds with Thymine from opposite strand and vice-versa. Guanine is bonded with Cytosine with three H-bonds. As a result, always a purine comes opposite to a pyrimidine. This generates approximately uniform distance between the two strands of the helix.
(iv) The plane of one base pair stacks over the other in double helix. This, in addition to H-bonds, confers stability to the helical structure.
(v) The two chains are coiled in a right-handed fashion. The pitch of the helix is 3.4 nm and there are roughly 10 bp in each turn. Consequently, the distance between base pairs in a helix is approximately equal to 0.34 nm. It is because of specific base pairing with a purine lying opposite to pyrimidine which makes two chains 2 nm thick.
Concept Builder
Types of DNA and their comparison
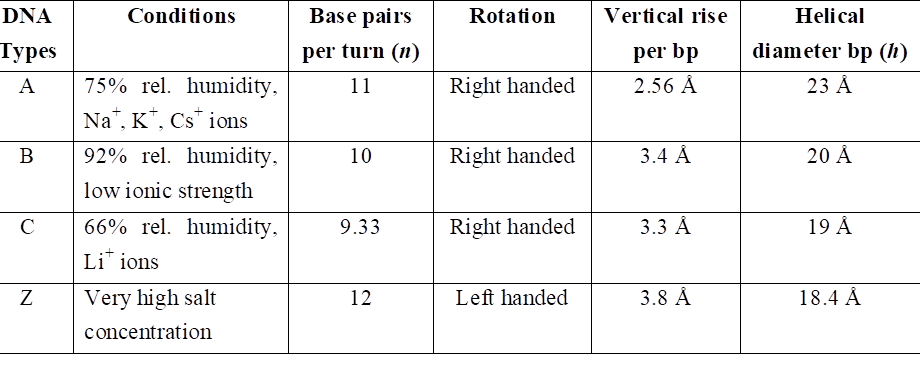
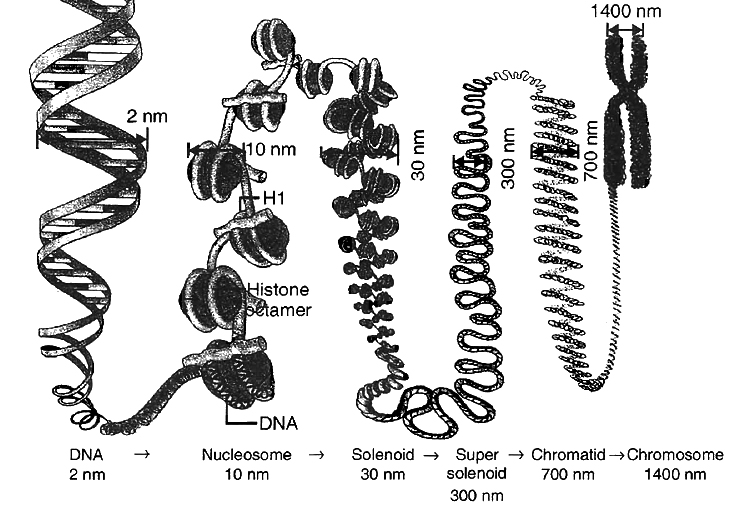
PACKAGING OF DNA HELIX
The average distance between two adjacent base pairs is 0.34 nm (0.34 × 10–9 m or 3.4 Å).
Length of DNA for a human diploid cell is 6.6 × 109 bp × 0.34 × 10–9 m/bp = 2.2 m.
This length is far greater than the dimension of a typical nucleus (approximately 10–6 m).
The number of base pairs in Escherichia coli is 4.6 × 106. The total length is 1.36 mm.
The long sized DNA is accommodated in small area (about 1 µm in E. coli) only through packing or compaction.
DNA is acidic due to presence of large number of phosphate group.
Compaction occurs by folding and attachment of DNA with basic proteins, polyamine in prokaryotes and histone in eukaryotes.
DNA packaging in Prokaryotes :
DNA is found in cytoplasm in supercoiled state.
The coils are maintained by non histone basic protein like polyamines.
RNA may also be involved. This compact structure of DNA is called nucleoid or genophore.
DNA packaging in Eukaryotes :
It is carried out with the help of lysine and ariginine rich basic proteins called histones.
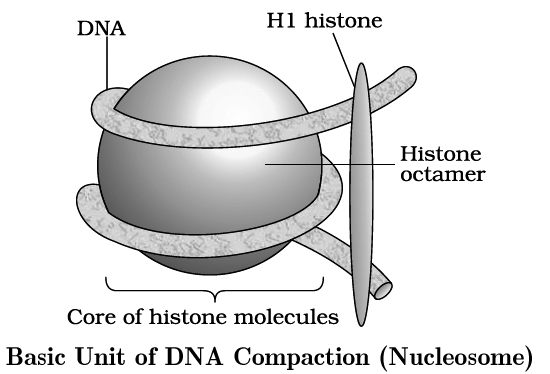
The unit of compaction is called nucleosome.
There are five types of histone proteins H1, H2A, H2B, H3 and H4.
Four of them occur in pairs to produce histone octamer (2 copies of each -H2A, H2B, H3 and H4), called nu body or core of nucleosome.
Their positively charged ends are directed outside.
They attract negatively charged strands of DNA.
About 200 bp of DNA is wrapped over nu body to complete about 1¾ turns.
This forms a nucleosome of size 110 × 60 Å (11 × 6 nm).
DNA present between two adjacent nucleosome is called linker DNA.
It is attached to H1 histone protein.
Length of linker DNA varies from species to species.
Nucleosome chain gives a beads on string appearance under electron microscope.
The nucleosomes further coils to form solenoid.
It has diameter of 30 nm as found in chromatin.
The beads-on-string structure in chromatin is packaged to form chromatin fibres that are further coiled and condensed at metaphase stage of cell division to form chromosomes.
The packaging at higher level requires additional set of proteins (acidic) that collectively are referred to as non-histone chromosomal (NHC) proteins.
Non-Histone Chromosomal proteins are of three types:
(i) Scaffold or Structural NHC protein.
(ii) Functional NHC protein, e.g., DNA polymerase, RNA polymerase.
(iii) Regulatory NHC protein, e.g., HMG (High mobility group proteins that controls gene expression).
In a typical nucleus, some region of chromatin are loosely packed (and stains light) and are referred to as euchromatin.
The chromatin that is more densely packed and stains dark is called as heterochromatin, specifically euchromatin is said to be transcriptionally active and heterochromatin is transcriptionally inactive.
Chemical Composition of Chromosome
A chromosome consists of following chemical compositions:
(a) DNA 40%
(b) RNA 1.2%
(c) Histone protein 50%
(d) Acidic proteins 8.5%
(e) Lipid traces
(f) Ca+2, Mg+2, Fe+2 traces

 ACME SMART PUBLICATION
ACME SMART PUBLICATION
