Mendel's Laws of Inheritance
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
INHERITANCE: HEREDITY AND VARIATIONS
Heredity :
It is the transmission of genetic characters from parents to the offsprings.
It deals with the phenomenon of "like begets like", e.g., human babies are like human beings in overall characteristics.
About 200 characters are found to be hereditary in man.
Variations are common in sexually reproducing organisms.
Asexually reproducing organisms are monoparental, hence exhibit no genetic variations.
Concept Builder
Variations are of following two types:
(1) Somatogenic Variations: These are acquired variations and are non-inheritable in nature. The ability of an organism to alter its phenotype in response to environment is called phenotypic plasticity.
(2) Blastogenic Variations: These are germinal variations and are hereditary in nature. They are again of two types :
(a) Continuous Variations: These are the fluctuating variations and can not give rise to new species. These are further of two types :
(i) Substantive: Variation in size, shape and colour of organism.
(ii) Meristic: Variation in number of parts, e.g., number of grains in an ear of wheat.
(b) Discontinuous Variations: Also known as mutations, sports or saltations. These variations are responsible for formation of new species and the organism thus formed, is called' a mutant.
Types of discontinuous variations :
(i) Substantive variations: These influence shape, colour, size etc., e.g., hairless cat, short legged ancon sheep.
(ii) Meristic variations: These affect number of parts, e.g., polydactyly in humans. Variations are of great significance in evolution as they make the organism better suited to modifying environmental conditions, produce new trait in organism and provide raw material for evolution.
Branches of Genetics
(i) Transmission genetics or Classical genetics, e.g., It is the study of Mendelian genetics and non-Mendelian genetics.
(ii) Forward genetics : It is the identification of mutated gene using the mutated phenotype.
(iii) Reverse genetics : It is the study of genes whose protein products are unknown.
(iv) Cytogenetics: It is the study of various aspects of chromosomes.
(v) Molecular/biochemical genetics : It is the study of structure and functions of genes.
(vi) Population or biometrical genetics : It is the study of the behaviour and effects of gene in population using mathematical models.
(vii) Behavioural genetics: It is the study of interaction of genes with the environment to produce a particular pattern of behaviour.
Pre-Mendelian Ideas About Inheritance or Theories of Blending Inheritance
The science of genetics arose with the rediscovery of Mendelism in 1900. Early philosophers, thinkers and workers have presented various theories to explain the phenomenon of inheritance.
These are called theories of blending inheritance. Some of these theories are:
(a) Moist Vapour Theory (Pythagoras : 500 B.C.) : Various body parts emit certain vapours, which get aggregated to form new individual.
(b) Reproductive Blood Theory (Aristotle: 384-322 B.C.) : According to him, menstrual fluid and semen are kinds of highly purified blood. Menstrual fluid provide inert substance for embryo formation and semen provide form and shape to embryo.
(c) Preformation Theory or Homunculus theory (J. Swammerdam) : According to this theory miniature form of individual is already present in sperm or egg called "homunculus". Fertilization is required to stimulate its growth.
(d) Theory of Pangenesis (Darwin 1868) : According to him, each part of body produces minute particle called gemmules or pangenes, which aggregate into gamete. On fusion they give rise to new individual.
(e) Theory of Epigenesis (K.F. Wolff) : According to this idea,· neither egg nor sperm had a structural homunculus but the gametes contained undifferentiated living substance capable of forming the organized body after fertilization. This suggested that many new organs and tissue which were originally absent, develop structurally de novo due to mysterious vital force.
Theory of pangenes was disproved by Weismann.
A.Weismann proposed his theory of germplasm, according to which the changes which affect the germplasm are heritable and the changes which affect somatoplasm are non heritable.
Objections to blending inheritance:
1. Unisexual traits
2. Skin colour in humans
3. Atavistic character
GENETIC TERMS AND SYMBOLS
MENDELIAN INHERITANCE
Mendel was born on July 22, 1822. He worke
d on Pisum sativum (Garden pea or Edible pea) for 7 years by taking 7 pairs of contrasting traits.
The results were read out in two meeting of Natural History Society of Brunn in 1865.
His paper "Experiments on plant Hybridisation" was published in 4th volume of "Proceedings of Natural Science Society of Brunn" in 1866.
Mendel was first to apply statistical analysis and mathematical logic.
He selected 14 true breeding pea plant varieties. He died due to kidney disorder in 1884.
Mendel selected following characters in pea plant for carrying out hybridisation experiments:

Mendel failed to produce same results in Hawkweed (Hieracium) and Beans (Lablab). Detailed investigation by S. Blixt on pea plant led to locate Mendel's seven characters on 4 different chromosomes, i.e., 1, 4, 5 and 7.
However, Mendel's work did not receive any recognition, it deserved, till 1900.
Mendel's work remained unnoticed and unappreciated for several years due to following reasons:
(1) Communication was not easy in those days and his work could not be widely publicised.
(2) His concept of stable, unblending, discrete units or factors for various traits did not find acceptance from the contemporaries.
(3) His approach of using mathematical and statistical analysis to explain biological phenomena was totally new and unacceptable to many of the biologists of that time.
(4) He could not provide any physical proof for the existence of factors. It was rediscovery of his work by a Dutch -Hugo de Vries, a German -Carl Correns and an Austrian botanist -Erich von Tschermak, independently in 1900, that brought Mendel to limelight. Correns raised status of Mendel's generalisations to laws.
Selection of pea plant : The main reasons for adopting garden pea (Pisum sativum) for experiments by Mendel were as follows :
(1) Pea has many distinct alternative traits (clear contrasting characters).
(2) Life span of pea plant is short.
(3) Flowers show self (bud) pollination, so are true breeding.
(4) It is easy to artificially cross-pollinate the pea flowers. The hybrids thus produced were fertile.
Concept Builder
1. Term allele was given by Bateson, term homozygous and heterozygous were given by Bateson and Saunders; genotype, phenotype, gene and pureline by Johannsen.
2. Father of genetics -Mendel; Father of Modem genetics -Bateson
3. Isoalleles : Alleles that produce similar phenotypes but are distinguishable amongst themselves through changed optima, e.g.,  .
.
4. Pseudoalleles : Genes are present together side by side and they produce related phenotypes. These are distinguished from true alleles through rare crossing over, e.g., star (dominant) and asteroid (recessive) traits in Drosophila.
5. Pure lines (pure breeding line) : A population obtained by continuous inbreeding over many generations, such that each individual has essentially the same genome as every other member of the inbred line and that all (or most) loci are homozygous.
Mendel's Work and Results
Mendel has made cross between parents having contrasting traits.
Firstly he made monohybrid cross (cross between parents, differ from each other in one character) followed by dihybrid cross (cross between parents, differ from each other in two characters) and then trihybrid.
The F1 hybrids were self crossed to give rise to F2 generation.
Mendel also carried out the reciprocal crosses and found that reciprocal crosses gave the same result.
(Reciprocal cross means opposite cross, i.e., the parent which provides male gamete in one cross, in second experiment it provides the female gamete and vice versa).
The result of reciprocal cross proves that both gametes produce the same effect and it does not matter which parent provides male and which one provides female gamete.
On the basis of his experimental crosses, he formulated four postulates.
Postulate I:
According to this postulate characters are controlled by a pair of unit factors. The two factors are now called alleles or allelomorphic pair.
Postulate II :
If two dissimilar unit factors are present in an individual, only one expresses itself. The one which expresses itself is known as dominant factor, while the second which does not express at all is known as recessive factor.
Postulate III :
According to this postulate, two contrasting alleles responsible for contrasting traits present in an individual do not get mixed and get separated from each other at the time of gamete formation by F1 hybrid and due to their recombination, four combinations can be obtained in equal frequency.
All the above three postulates are based upon Mendel's monohybrid cross or one gene interaction.
Law of dominance and law of segregation can be explained on the basis of monohybrid cross or one gene interaction.
(1) Law of dominance :
This law states that when two contrasting alleles for a character come together in an organism, only one is expressed completely and shows visible effect.
It is called dominant and the other allele of the pair which does not express and remains hidden is called recessive.
This law is not universally applicable.
Plant height is controlled by two alleles -Dominant allele (T) and Recessive allele (t)
These two alleles can be present in three forms

Tt -Dominant allele expressed] Heterozygous (Hybrid for character)
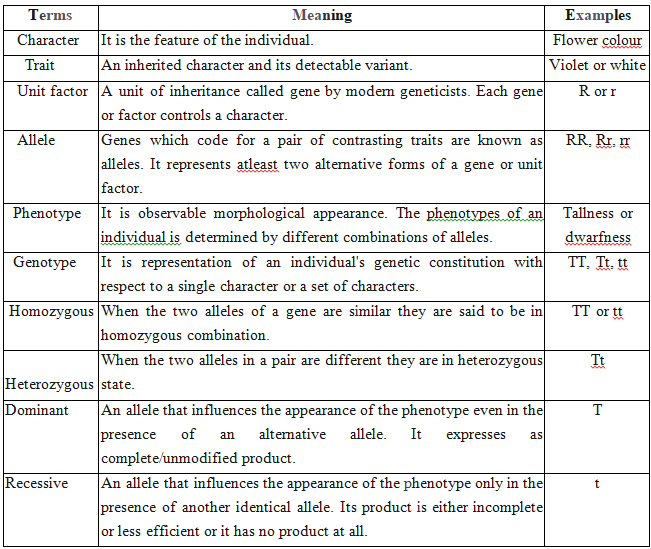
Mendel crossed two pea plants, one homozygous tall (TT) and another homozygous dwarf (tt).
He observed that all the F1 progeny plants were tall, like one of the parents, none were dwarf.
He made similar observations for the other pair of traits and found that F1 always resembled only one of the parents, and that the trait of other parent was not seen in them.
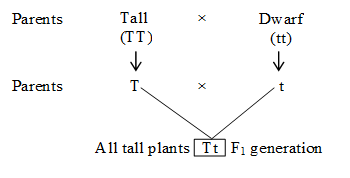
(2) Law of segregation or Law of purity of gametes:
This law states that both parental alleles (recessive and dominant) of F1 separate and are expressed phenotypically in F2 generation. This law is universally applicable.
The F2 generation was produced by allowing F1 hybrid to self pollinate, to find out segregation or separation.
It was observed that both dominant and recessive plants appeared in 3 : 1 ratio. Thus, F2 progeny shows both parental forms.
On the basis of F2 generation, following observations can be made:
(i) An organism generally has two alleles for each character. These alleles may either be similar or dissimilar. Organism with similar alleles of a pair is called pure or true breeding for that character. If the organism contains dissimilar alleles of a pair, the organism is impure or hybrid.
(ii) An organism receives one of the two alleles from the male gamete and the other from the female gamete. The gametes fuse during fertilization and form a zygote. Zygote develops into an organism.
(iii) Each gamete (male or female) has only one allele of the pair. Thus, each gamete is pure for a trait. That is why this law is often called as Law of purity of gametes.
(iv) The fusion between male and female gametes to produce zygote is a random process.
The plants obtained in F2 generation show 3 (tall) : 1 (dwarf) phenotypic ratio. Of these three tall plants, one is pure or homozygous dominant and the remaining two are heterozygous (tall in this case). There is only one plant that shows recessive character (dwarf in this case). Dwarf is pure or true breeding, being homozygous recessive.
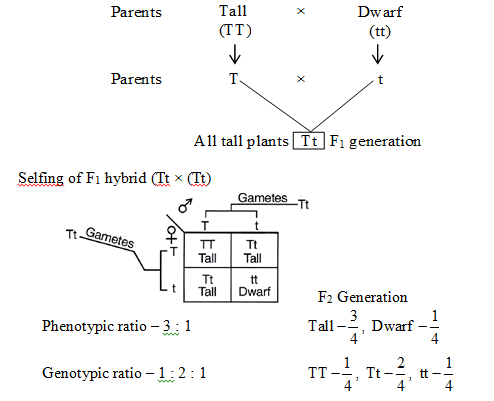
Postulate IV :
This postulate was made on the basis of dihybrid cross or two genes interaction.
He postulated that inheritance of one character is independent of the inheritance of another character.
On the basis of this postulate, Mendel proposed the "Law of independent assortment".
(3) Law of independent assortment :
The law of independent assortment states that when a cross is made between two individuals different from each other in two or more characters, then the inheritance of one character is independent of the inheritance of another character.
Because of their independent assortment, besides the parental types, recombinants are also obtained.
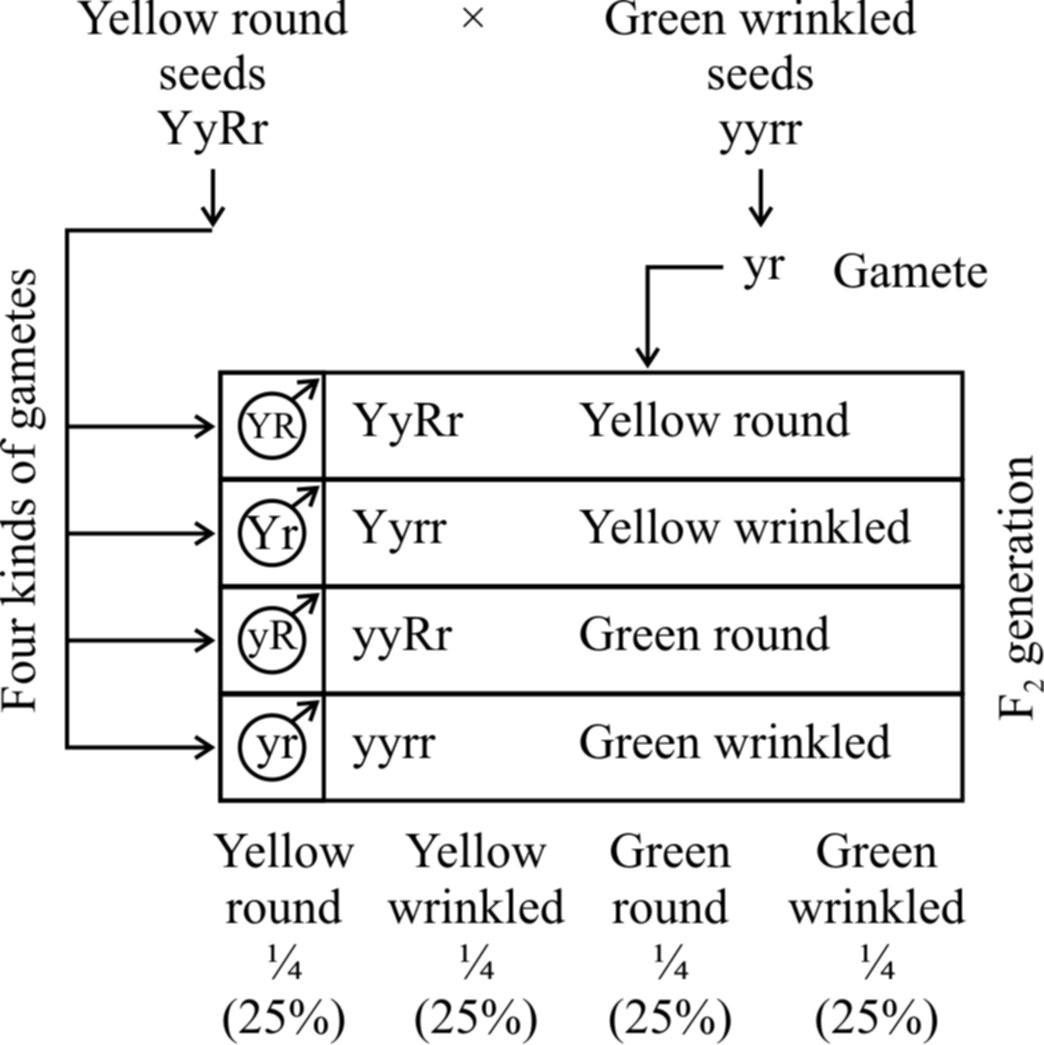
In dihybrid cross, these combinations are obtained in the ratio of 9 : 3 : 3 : 1. e.g., He crossed homozygous dominant round and yellow seeded plant (RRYY) with homozygous recessive wrinkled and green seeded (rryy) plant.
The F1 hybrids were all heterozygous, showing yellow and round seeded plants. This law is not universally applicable.
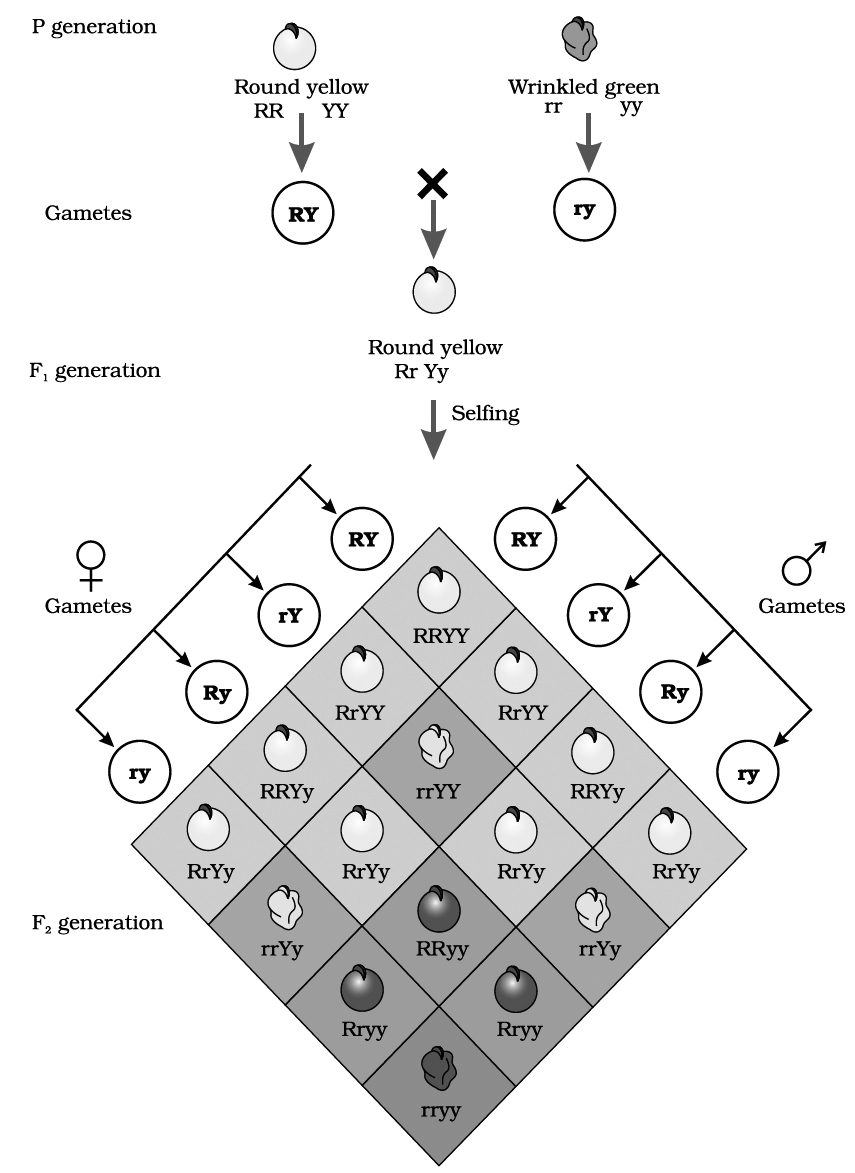
two pairs of contrasting traits : seed colour and seed shape
Phenotypic ratio: round yellow : round green : wrinkled yellow : wrinkled green
9/16 3/16 3/16 1/16
Genotypic ratio : 1 : 2 : 1 : 2 : 4 : 2 : 1 : 2 : 1
If the phenotypic ratio of each pair of alleles (e.g., yellow and green colour of seed) is considered, it shows 12(9 + 3) yellow seeded plants and 4(3 +1) green seeded plants.
This comes to 3 : 1 ratio; similar to one obtained in F2 generation of monohybrid cross showing segregation.
The same is true for another pair of alleles involved, i.e., round and wrinkled seeded plants. So, the results of each character are similar to the monohybrid ratio.
SUMMARISED ACCOUNT OF MENDEL'S EXPERIMENTS:
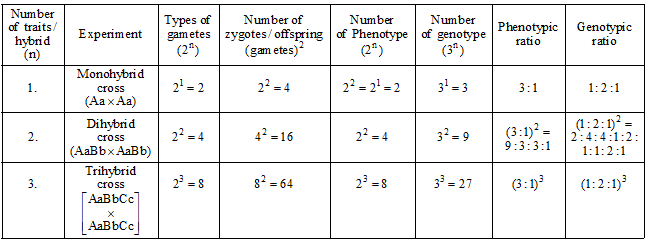
Back Cross and Test Cross
Back cross:
F1 hybrids are obtained by crossing two plants of parental generation.
Mendel devised a cross where F1 hybrid is crossed with anyone of the two parents, i.e., homozygous dominant and homozygous recessive.
Thus, there would be two possibilities:
(a) F1 hybrid (Tt) is crossed with homozygous dominant (TT)
(b) F1 hybrid (Tt) is crossed with homozygous recessive (tt)
Both these crosses collectively are called as back cross. If F1 is crossed with dominant parent, it is called out cross.
Test Cross:
Out of the two types of back crosses, a cross between F1 hybrid (Tt) and its homozygous recessive parent (tt) is called test cross.
This cross is called test cross because it helps to find out whether the given dominant F1 phenotype is homozygous or heterozygous.
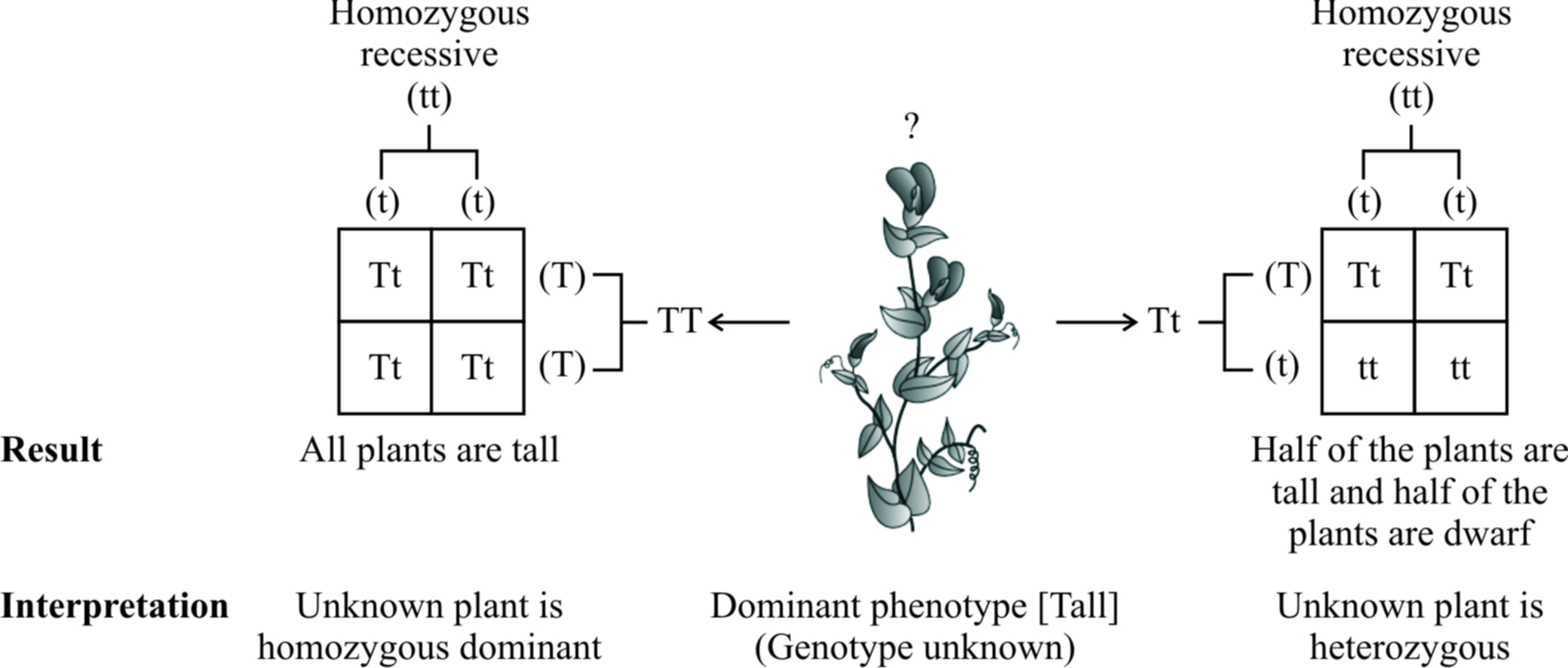
A monohybrid test cross between F1 tall plant (Tt) and its homozygous recessive parent (tt) will produce 50% heterozygous tall (Tt) and 50% homozygous recessive (tt), i.e., 1 : 1 ratio, for both phenotype and genotype.
If a test cross with two characters, i.e., dihybrid test cross is made, it gives four types of plants in 1 : 1 : 1 : 1 ratio.
The phenotypes obtained are similar to those found in F2 generation of dihybrid cross.
Thus, a dihybrid test cross between F1 yellow and round seeded plant (YyRr) and its homozygous recessive parent green and wrinkled (yyrr) would give following combinations:
1 yellow, round (YyRr) Parental combination 25%
1 yellow, wrinkled (Yyrr) Recombinants 25%
1 green, round (yyRr) Recombinants 25%
1 green, wrinkled (yyrr) Parental combination 25%
If this ratio is obtained, it would be confirmed that F1 hybrid with dominant phenotype is infact heterozygous.
The parental combinations (50%) are equal to the frequency of recombinants (50%).
Trihybrid cross.
Mendel crossed two pea plants which differed in 3 characters and observed independent assortment of genes in them.
He crossed two pea plants pure in three traits viz., height of stem, form of seed and colour of cotyledon of seed.
He crossed homozygous tall, round and yellow (TT RR YY) plant with dwarf, wrinkled and green (tt rr yy).
All the F1 individuals produced were tall, round and yellow (Tt Rr Yy) and are called trihybrids.
On selfing trihybrids, F2 phenotypic ratio is 27 : 9 : 9 : 9 : 3 : 3 : 3 : 1. The ratio for a trihybrid test cross is 1 : 1 : 1 : 1 : 1 : 1 : 1 : 1 .
Inheritance of one gene
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
ONE GENE INTERACTION (w.r.t. Post-Mendelian inheritance)
(1) Incomplete dominance:
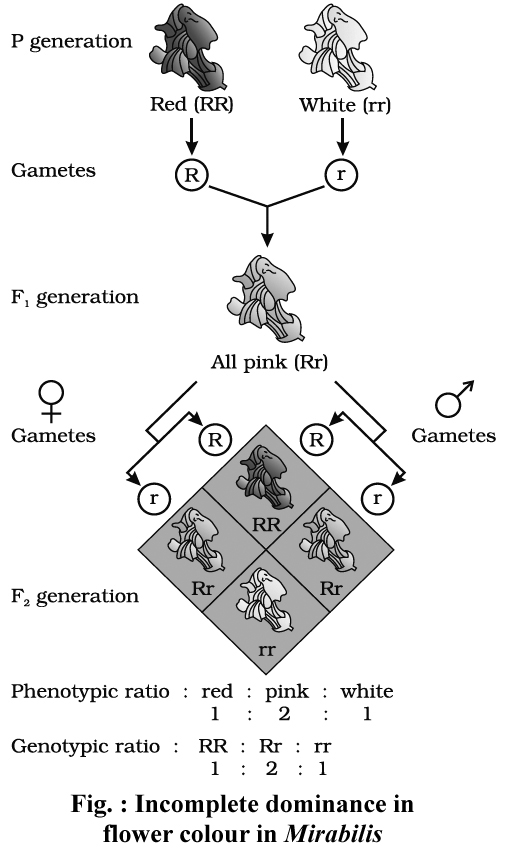
After Mendelism a few cases were observed where F1 phenotype is intermediate between dominant and recessive phenotypes.
The most common example of incomplete dominance is that of flower colour in Mirabilis jalapa (Gulbansi or 4'O clock plant), studied by Carl Correns.
Homozygous red (RR) flowered variety was crossed with white (rr) flowered variety.
F1 offspring had pink flowers.
Thus, here one allele is incompletely dominant over the other so that intermediate phenotype is produced by F1 hybrid with respect to the parents.
This is called incomplete dominance.
Incomplete dominance for flower colour [Red(RR), Pink(Rr), White (rr)] is also known to occur in Antirrhinum majus (Snapdragon or Dog flower).
The phenotypic ratio and genotypic ratio in F2 generation is identical in case of incomplete dominance i.e., 1 : 2 : 1.
Explanation of the concept of dominance:
Every gene contains information to express a particular trait.
Diploid organisms have two copies of each gene, they are called alleles.
These two alleles may be identical or non-identical.
One of them may be different due to some changes that it has undergone which modifies the information that particular allele contains.
Theoretically, the modified allele could be responsible for production of
(i) The normal/less efficient enzyme, or
(ii) a non-functional enzyme, or
(iii) no enzyme at all
In case (i), the modified allele is equivalent to the unmodified allele, i.e., it will produce the same phenotype/trait.
But, if the allele produces a non-functional enzyme or no enzyme [case (ii) &(iii)], the phenotype may be effected.
The unmodified (functioning) allele, which represents the original phenotype is the dominant allele and the modified allele is generally the recessive allele.
Hence, the recessive trait is due to nonfunctional enzyme or because no enzyme is produced.
If the mutated allele forms an altered but functional product, it behaves as incompletely or codominant allele.
(2) Multiple allelism:
Mendel proposed that each gene has two contrasting forms, i.e., alleles.
But there are some genes which are having more than two alternative forms (allele).
Presence of more than two alleles for a gene is known as multiple allelism.
Multiple alleles are present on the same locus of homologous chromosome.
Multiple alleles can be detected only in a population.
A well known example to explain multiple alleles in human beings is ABO blood type.
Landsteiner discovered ABO system of blood groups. The fourth group AB was discovered by de Castello and Steini.
Bernstein showed that these groups are controlled by 3 alleles -IA, IB and IO/i.
These alleles are autosomal and follow Mendelian pattern of inheritance.
The alleles IA and IB produce a slightly different form of the sugar while IO doesn't produce any sugar. Because humans are diploid organism, each person possesses any two of the three I gene alleles.
IA and IB are completely dominant over IO, but when IA and IB are present together they both express their own types of sugar thus, behaving as codominant alleles.
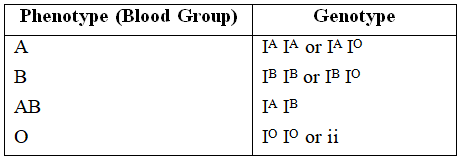
Possible blood types of children from parents of various blood types.

Other examples of multiple alleles are - coat colour in rabbit, eye colour in Drosophila and self incompatibility in tobacco. Formula to find out number of genotypes for multiple allelism is n/2(n +1) , where 'n' is number of alleles.
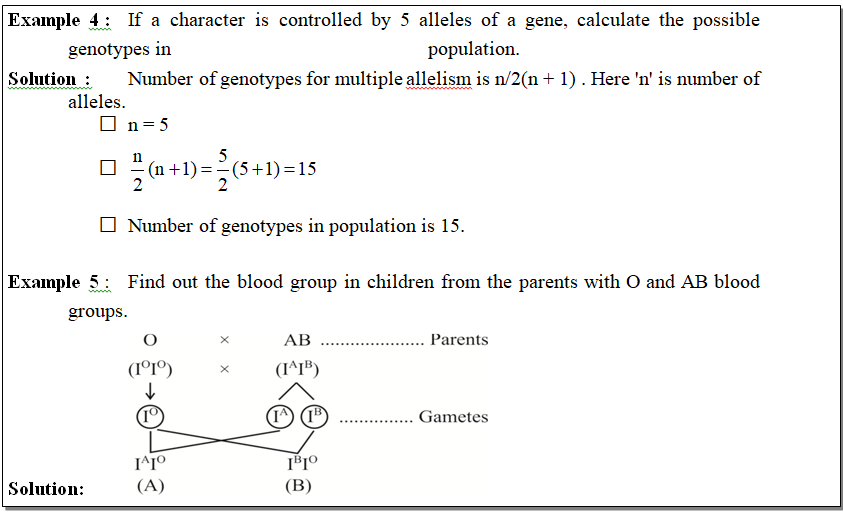
The possibility of blood groups in offsprings of this couple is A and B.
(3) Co-dominance.
In co-dominance, the genes of an allelomorphic pair are not related as dominant and recessive but both of them express themselves equally in F1 hybrids.
These follow the law of segregation and F2 progeny exhibits 1 : 2 : 1 ratio. Heterozygous for sickle cell anaemia (HbAHbS), AB and MN blood groups are examples of co-dominance of alleles.
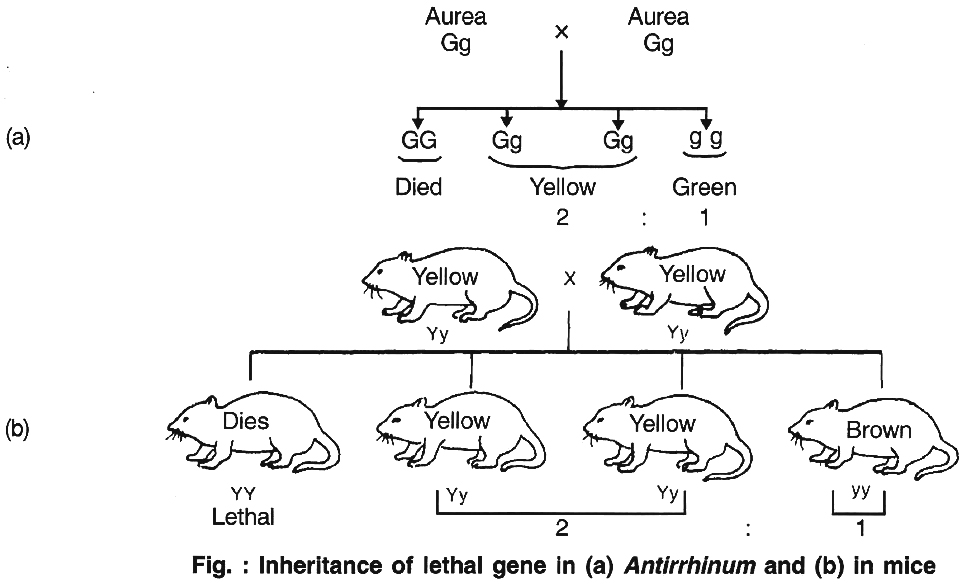
(4) Lethal genes or lethality.
A lethal gene usually results in the death of an individual when present in homozygous condition. Most striking example to explain lethal gene is sickle cell anaemia (HbSHbS).
Cuenot (1905) first reported that inheritance in mouse body colour did not agree with Mendelian inheritance, because the dominant allele for yellow body colour is lethal in homozygous condition.
The homozygous dominant gene carrying mouse died, proving dominant gene is lethal in homozygous form.
This is called absolute lethality. In plants, it was first reported in Antirrhinum majus by E. Baur, where yellow leaved or golden leaved or Aurea plant never breed true. Thus, the ratio comes out to be 2 : 1.
(5) Pleiotropic genes:
The ability of a gene to have multiple phenotypic effects (as it influences a number of characters simultaneously) is known as pleiotropy.
The gene having multiple phenotypic effects is called pleiotropic gene.
It is not essential that the traits are equally influenced. Sometimes, the effect of the gene is more evident in case of one trait (major effect) and less evident in case of others (secondary effect).
Occasionally, a number of related changes are caused by a gene.
They are together called syndrome.
Some common examples in humans are -Cystic fibrosis, Marfan syndrome and Phenylketonuria, while in Drosophila, a single gene influences the size of wings, character of balancers, position of dorsal bristles, eye colour, shape of spermathecae, fertility and longevity.
In human beings pleiotropy is exhibited by sickle cell anaemia in heterozygous condition (HbAHbS).
In case of pea, the gene which controls the starch synthesis also controls the shape of the seed.
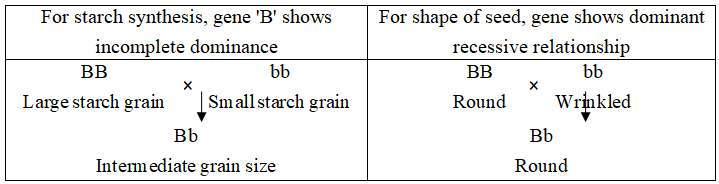
Concept Builder
(1) Rh factor: It is an antigen protein on RBC surface, reported by Landsteiner and Wiener in blood of Indian brown Rhesus monkey (Macacus rhesus). 93% Indians have this antigen and are called Rh positive (Rh+).
(2) Marian syndrome: Slender body, elongation of limbs, hypermobility of joints, dislocation of lens and susceptibility to heart diseases.
Inheritance of two genes
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
TWO GENES INTERACTION (w.r.t. Post-Mendelism)
Genes usually function or express themselves singly or individually.
But, many cases are known where two genes of the same allelic pair or genes of two or more different allelic pairs influence one another.
This is called gene interaction.
Non-allelic genetic interactions.
These are interactions between genes located on the same chromosome or on different but non-homologous chromosomes controlling a single phenotype to produce a different expression.
Each interaction is typical in itself and ratios obtained are different from those of the Mendelian dihybrid ratios.
Some of these interactions of genes are explained here which fall under this category and deviate from Mendel's ratios.
1. Complementary genes :
The complementary genes are two genes present on separate loci that interact together to produce dominant phenotypic character, neither of them if present alone, can express itself. It means that these genes are complementary to each other.
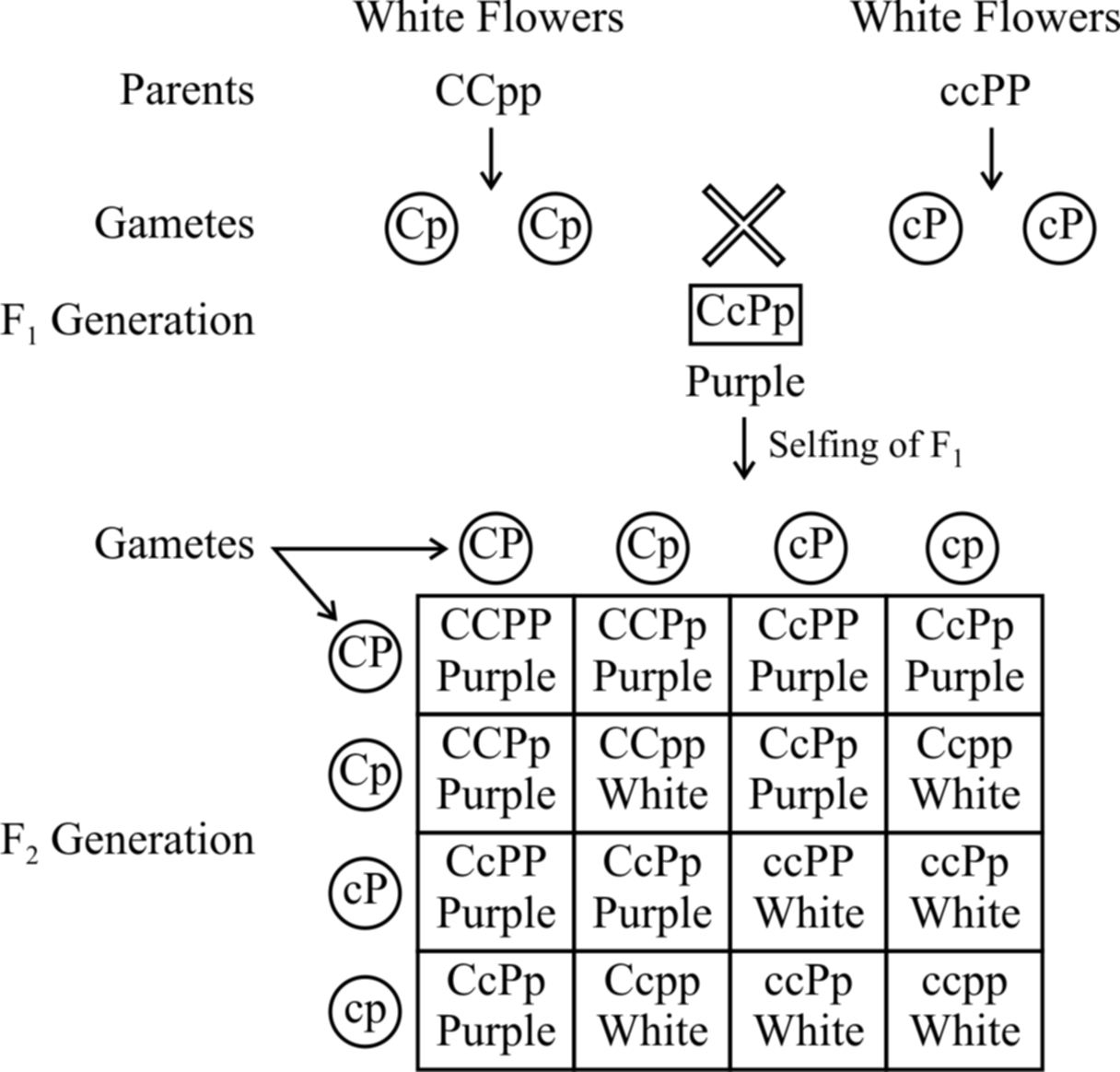
Fig. : Results of an experiment showing inheritance of flower colour in
Lathyrus odoratus controlled by complementary genes
Bateson and Punnet have demonstrated that in sweet pea (Lathyrus odoratus) purple colour of flowers develop as a result of interaction of two dominant genes C and P.
In the absence of dominant gene C or P or both, the flowers are white.
It is believed that gene C produces an enzyme that catalyzes the formation of necessary raw material for the synthesis of pigment anthocyanin and gene P produces an enzyme which transforms the raw material into the pigment.
It means the pigment anthocyanin is the product of two biochemical reactions, the end product of one reaction forms the substrate for the other.
![]()
Therefore, if a plant has ccPP, ccPp, CCpp or Ccpp genotypes, it bears only white flowers. Purple flowers are formed in plants having genotype CCPP or CCPp or CcPP or CcPp.
From checker board, it is clear that 9 : 7 ratio between purple and white is a modification of 9 : 3 : 3 : 1 ratio.
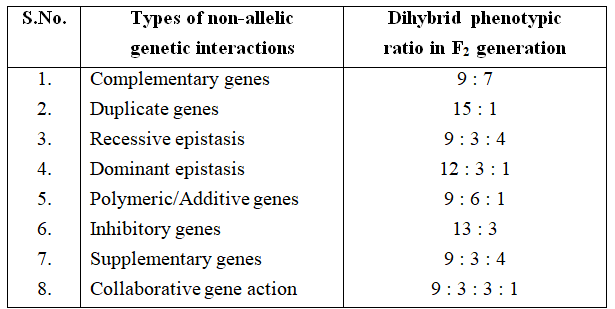
2. Duplicate genes.
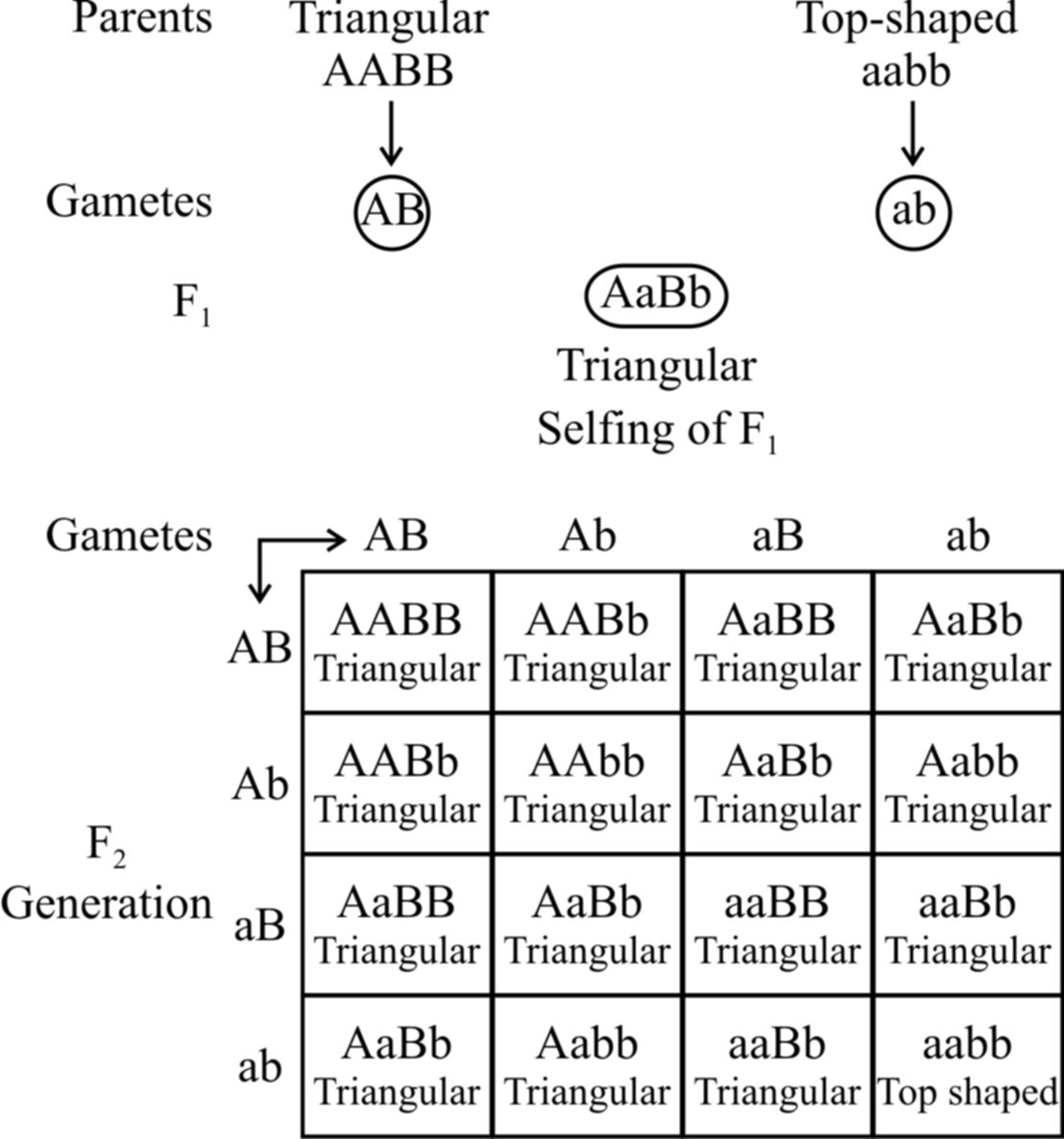
If the dominant alleles of two gene loci produce the same phenotype, whether inherited together or separately, the 9 : 3 : 3 : 1 ratio is modified into a 15 : 1 ratio.
Example: The capsules of shepherd's purse (Capsella) occur in two different shapes, i.e., triangular and top-shaped. When a plant with triangular capsule is crossed with one having top-shaped capsule, in F1 only triangular character appears. The F1 offspring by self crossing produced the F2 generation with the triangular and top-shaped capsules in the ratio of 15 : 1. Two independently segregating dominant genes (A and B) have been found to influence the shape of capsule in the same way. All genotypes having dominant alleles of both or either of these genes (A and B) would produce plants with triangular-shaped capsules. Only those with the genotype aabb would produce plants with top shaped capsules. The results of this example are given below.
3. Epistasis
A gene which masks (hides) the action of another gene (non allelic) is termed as epistatic gene. The process is called epistasis. The gene whose effects are masked is called hypostatic gene.
Epistasis is of two types :
(a) Recessive epistasis :
Here the recessive allele in homozygous condition masks the effect of dominant allele, e.g., in mice, the wild body colour is known as agouti (greyish) and is controlled by a gene say A which is hypostatic to recessive allele c.
The dominant allele C in the presence of 'a' gives coloured mice.
In the presence of dominant allele C, A gives rise to agouti.
So, CCaa will be coloured and ccAA will be albino.
When coloured mice (CCaa) are crossed with albino (ccAA), agouti mice (CcAa) appear in F1.
cc masks the effect of AA and is therefore epistatic.
Consequently, cc AA is albino.
The ratio 9 : 3 : 3 : 1 is modified to 9 : 3 : 4.
The combination ccaa is also albino due to the absence of both the dominant alleles.
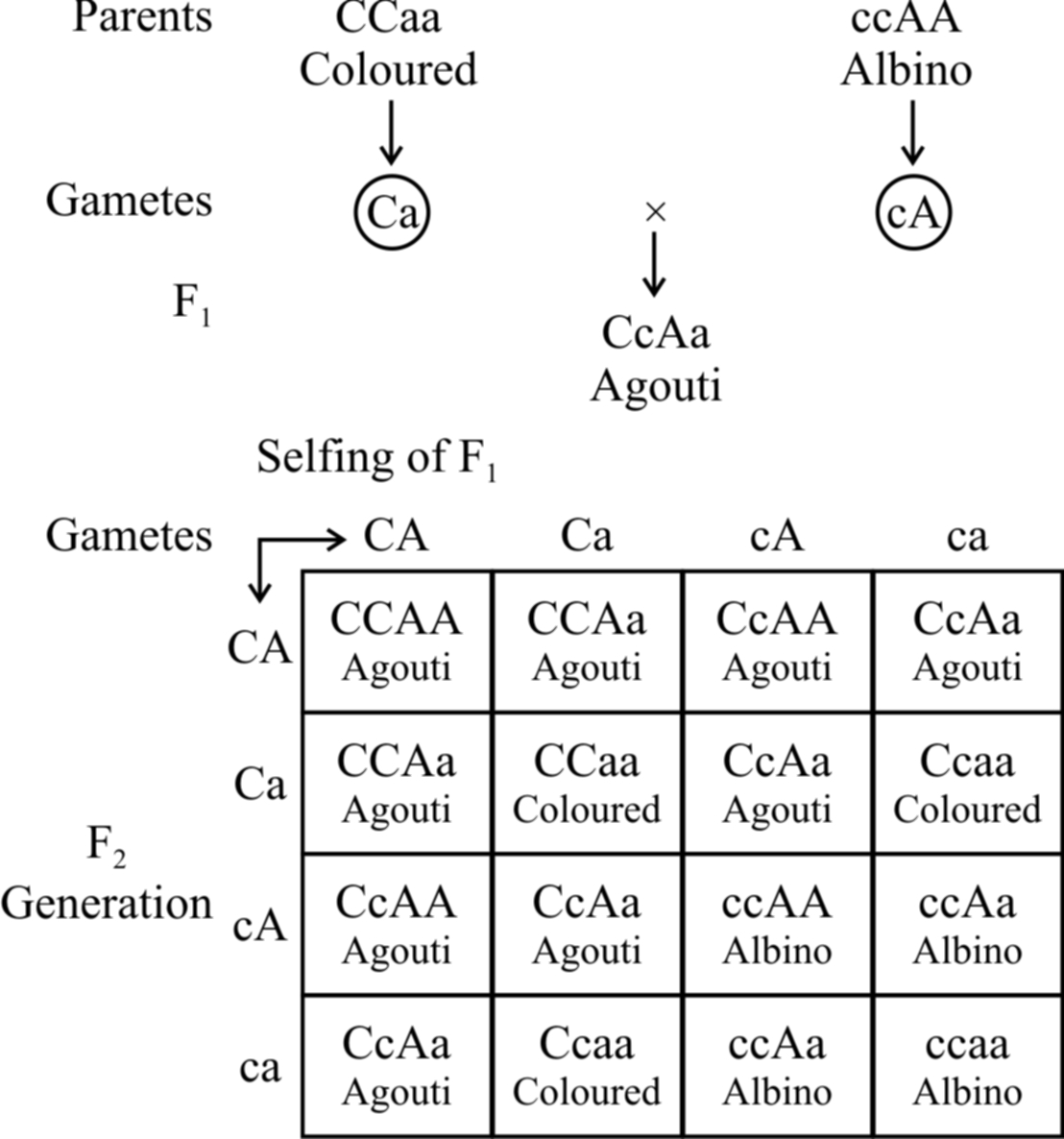
Fig. : Recessive epistasis
(b) Dominant epistasis :
In summer squash or Cucurbita pepo, there are three types of fruit colour -yellow, green and white.
White colour is dominant over other colours, while yellow is dominant over green.
Gene for white colour (W) masks the effects of yellow colour gene (Y).
So, yellow colour is formed only when the dominant epistatic gene is represented by its recessive allele (w). When the hypostatic gene is also recessive (y) , the colour of the fruit is green.
White Fruit – W – Y –, W – y –
Yellow Fruit – wwY –
Green Fruit – wwyy
A cross between a pure breeding white summer squash, (WWYY) with a pure breeding green summer squash, (wwyy) yields white fruits in the F1 generation. Upon selling of F1, the F2 generation comes to have 12 white fruit : 3 yellow fruit : 1 green fruit.
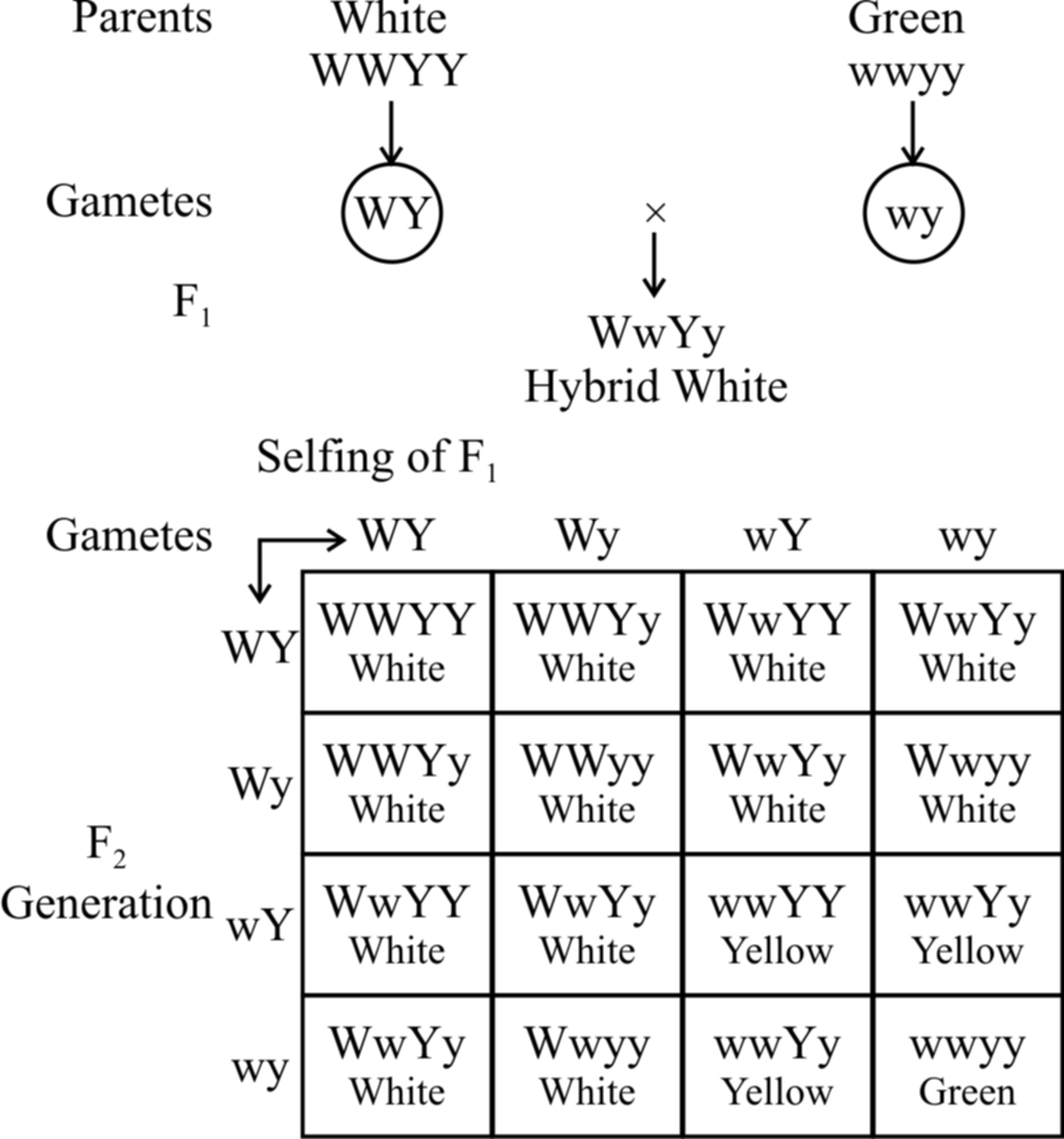
Fig. : Dominant epistasis
Polygenic inheritance or quantitative inheritance
Quantitative inheritance is controlled by two or more genes in which the dominant alleles have cumulative effect, with each dominant allele expressing a part of functional polypeptide and full trait is shown when all the dominant alleles are present.
Genes involved in quantitative inheritance are called polygenes.
H. Nilsson-Ehle (1908) and East (1910) demonstrated segregation and assortment of genes controlling quantitative traits, e.g., Kernel colour in wheat and corolla length in tobacco.
Kernel colour in wheat. Swedish geneticist, H. Nilsson-Ehle (1908) crossed red kerneled variety with white kerneled variety of wheat.
Grains of F1 were uniformly red but intermediate between the red and white of parental generation.
When members of F1 were self·crossed among themselves, five different phenotypic classes appeared in F2 showing the ratio of 1 : 4 : 6 : 4 : 1.
(i) The extreme red -1/16 (as red as to the parent of F1)
(ii) Deep red (Dark red) -4/16
(iii) Intermediate red -6/16 (similar to F1)
(iv) Light red -4/16
(v) White -1/16 (as white as to the parent of F1)
Nilsson Ehle found that the kernel colour in wheat is determined by two pairs of genes AA and BB.
Gene A and B determine the red colour of kernel and are dominant over their recessive alleles. Each gene pair shows Mendelian segregation.
Heterozygotes for two pairs of genes (AaBb) segregate into 15 red and one white kerneled plants.
But all the red kernels do not exhibit the same shade of redness.
The degree of redness was found to correspond with the number of dominant alleles.
Skin Colour in Man
The presence of melanin pigment in the skin determines the skin colour.
The amount of melanin developing in the individual is determined by three (two also) pairs of genes.
These genes are present at three different loci and each dominant gene is responsible for the synthesis of fixed amount of melanin.
The effect of all the genes is additive and the amount of melanin produced is always proportional to the number of dominant genes.
Subsequent studies after Davenport have shown that as many as six genes may be involved in controlling the skin colour in human beings.
As shown in table, the effect of all the genes is additive (The character is assumed to be fixed by three pairs of polygenes).
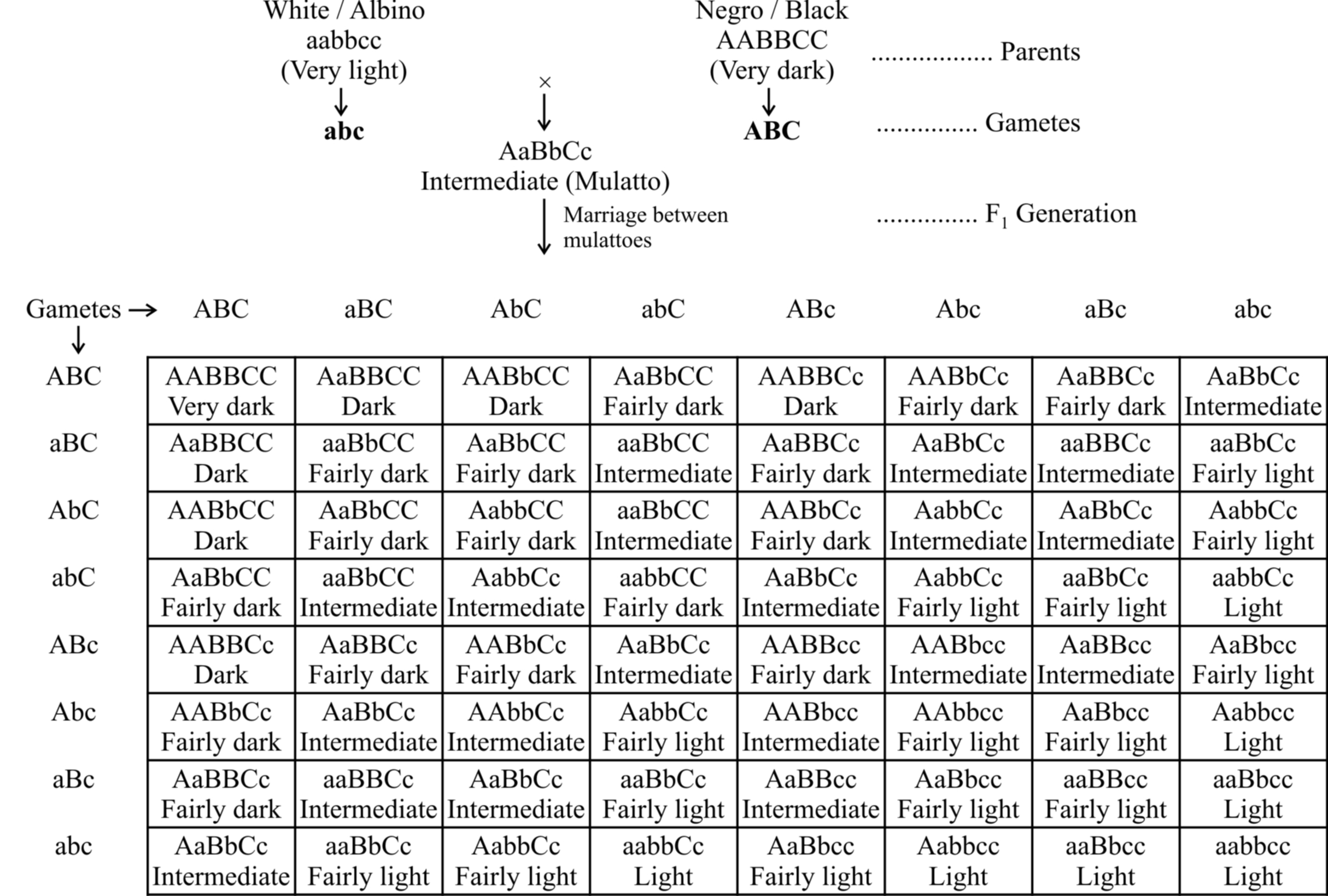
Phenotypes: 1 (Very dark) : 6 (Dark) : 15 (Fairly dark) : 20 (Intermediate) : 15 (Fairly light) : 6 (Light) : 1 (Very light)
The F1 progeny between an albino and a negro individual called mulatto produces intermediate skin colour.
In F2 generation, the coloured offsprings exhibit different shades in the ratio 1 : 6 : 15 : 20 : 15 : 6 : 1.
The frequency distribution for skin colour can be represented either as a histogram or in the form of a bell-shaped normal distribution curve.
Looking at the histogram, it can be concluded that in polygenic inheritance, the extreme phenotypes are rare and the intermediate ones are more frequent.
Some other example of quantitative traits are cob length in maize, human intelligence, milk and meat production, height in human and size, shape and number of seeds and fruits in plants.
- Number of phenotype for polygenes = 2n + 1
- Number of genotype for polygenes = 3n, where n represent pair of polygenes.
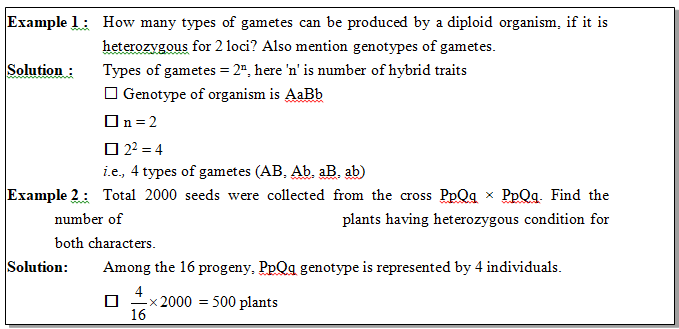
Example.7 : Calculate the sum total of phenotypes and genotypes in F2 generation if a character is controlled by 2 pair of polygenes.
Solution : Number of phenotype for polygenes = 2n + 1
(2 × 2 + 1) = 5
Number of genotype for polygenes = 3n
32 = 9
i.e., 5 + 9 =14
CHROMOSOMAL THEORY OF INHERITANCE / PARALLELISM BETWEEN
CHROMOSOMES AND MENDELIAN FACTORS
Chromosomal theory of inheritance was proposed independently by Sutton and Boveri.
The two workers found a close similarity between the transmission of hereditary traits and behaviour of chromosomes while passing from one generation to the next through the agency of gametes.
Sutton and Boveri noted that the behaviour of chromosomes is parallel to the behaviour of Mendelian factors (Genes).
The salient features of chromosomal theory of inheritance are as follows :
1. Like the hereditary traits the chromosomes retain their number, structure and individuality throughout the life of an organism and from generation to generation. The two neither get lost nor mixed up. They behave as units.
2. Both chromosomes as well as genes occur in pairs in the somatic or diploid cells. The two alleles of a gene pair are located on homologous sites on homologous chromosomes.
3. A gamete contains only one chromosome of a type and only one of the two alleles of a trait.
4. The paired condition of both chromosomes as well as Mendelian factors is restored during fertilization.
Homologous chromosomes synapse during meiosis and then separate or segregate independently into different cells which establishes the quantitative basis for segregation and independent assortment of hereditary factors.
Sutton united the knowledge of chromosomal segregation with Mendelian principles and called it the chromosomal theory of inheritance.
Johannsen (1909) coined the term gene, for mendelian factor.
Following the synthesis of ideas, experimental verification of the chromosomal theory of inheritance by T.H. Morgan and his colleagues, led to discovery of the basis for the variations, that sexual reproduction produced.
Thomas Hunt Morgan (1866-1945) is known as father of experimental genetics. He was awarded Nobel Prize of physiology in 1933 for his pioneer work in experimental genetics.
Drosophila melanogaster as material for experimental Genetics
Fruit fly Drosophila is a tiny fly of about 2 mm size which is found over ripe fruits like mango and banana.
The fly is actually attracted to yeast cells present on the surface of the ripe fruits. Drosophila is more suitable than pea as experimental material because of following reasons :
(i) It can be easily reared and bred under laboratory conditions.
(ii) The fly has a short life span of about two weeks. The fruit fly can be bred throughout the year so that numerous generations can be obtained in a single year instead of one as in case of Pea.
(iii) A single mating produces hundreds of offsprings.
(iv) Females are easily distinguishable from the males by the larger body size and presence of ovipositor (egg laying structure).
(v) The animal shows a number of externally visible and easily identifiable contrasting traits.
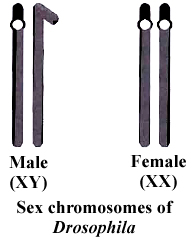
(vi) It has a smaller number (4 pairs) of morphologically distinct chromosomes.
(vii) Polytene chromosomes occur in the salivary glands of larva. Polytene chromosomes can be used to study different types of chromosome aberrations.
(viii) It has heteromorphic (XY) sex chromosomes in the male. The transmission of heteromorphic chromosomes can be easily studied from one generation to another.
LINKAGE (Exception to Law of Independent Assortment)
According to Mendel's law of independent assortment, the gene controlling different characters get assorted independent to each other.
It is correct if the genes are present on two different chromosomes, but if these genes are present on same chromosome they may or may not show independent assortment.
If crossing over takes place between these two genes then the genes get segregated and they will assort independent to each other. But if there is no crossing over between these two genes there is no segregation, hence only parental combination will be found in gametes.
The tendency of some of the genes to inherit together (in block) is known as linkage.
In 1906, Bateson and Punnet crossed two varieties of Lathyrus odoratus (sweet pea) and observed that the results do not agree with the Mendel's law of independent assortment.
They formulated the hypothesis of coupling and repulsion to explain the unexpected F2 results of dihybrid cross between a homozygous sweet pea having dominant alleles for blue flowers (BB) and long pollen grains (LL) with another homozygous double recessive plant with red flowers and round pollen grains (bbll).
Test cross ratio of 7 : 1 : 1 : 7 indicated that there was a tendency of the dominant alleles to remain together. Similar was the case with recessive alleles.
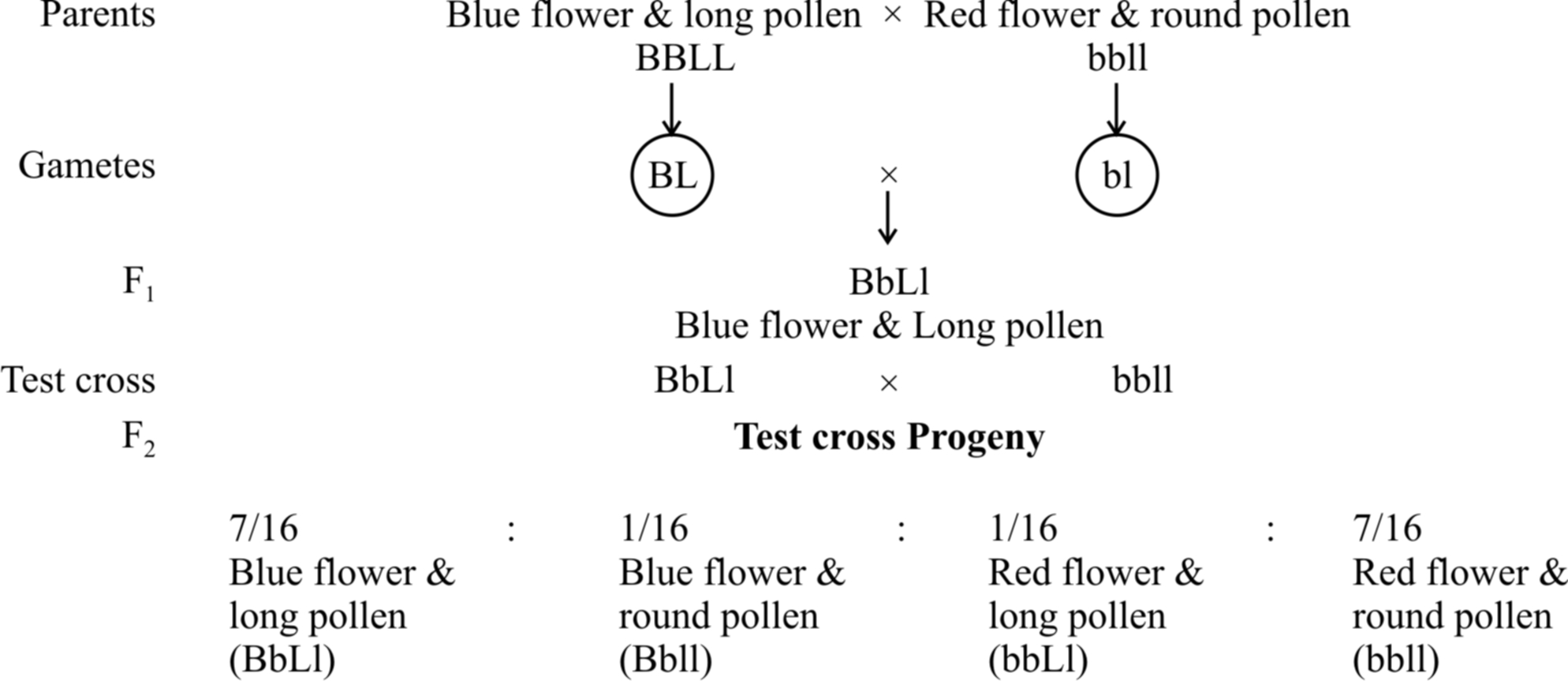
It was called gametic coupling by Bateson and Punnet.
Two dominant genes from one parent entered the same zygote more frequently than expected.
The tendency of two dominant genes to remain together in the process of inheritance was called as coupling.
In another cross, they took a sweet pea plant with blue flowers and round pollens (BBII) and other plant with red flowers and long pollens (bbLL) and obtained the ratio of 1 : 7 : 7 : 1 by test crossing F1 generation.
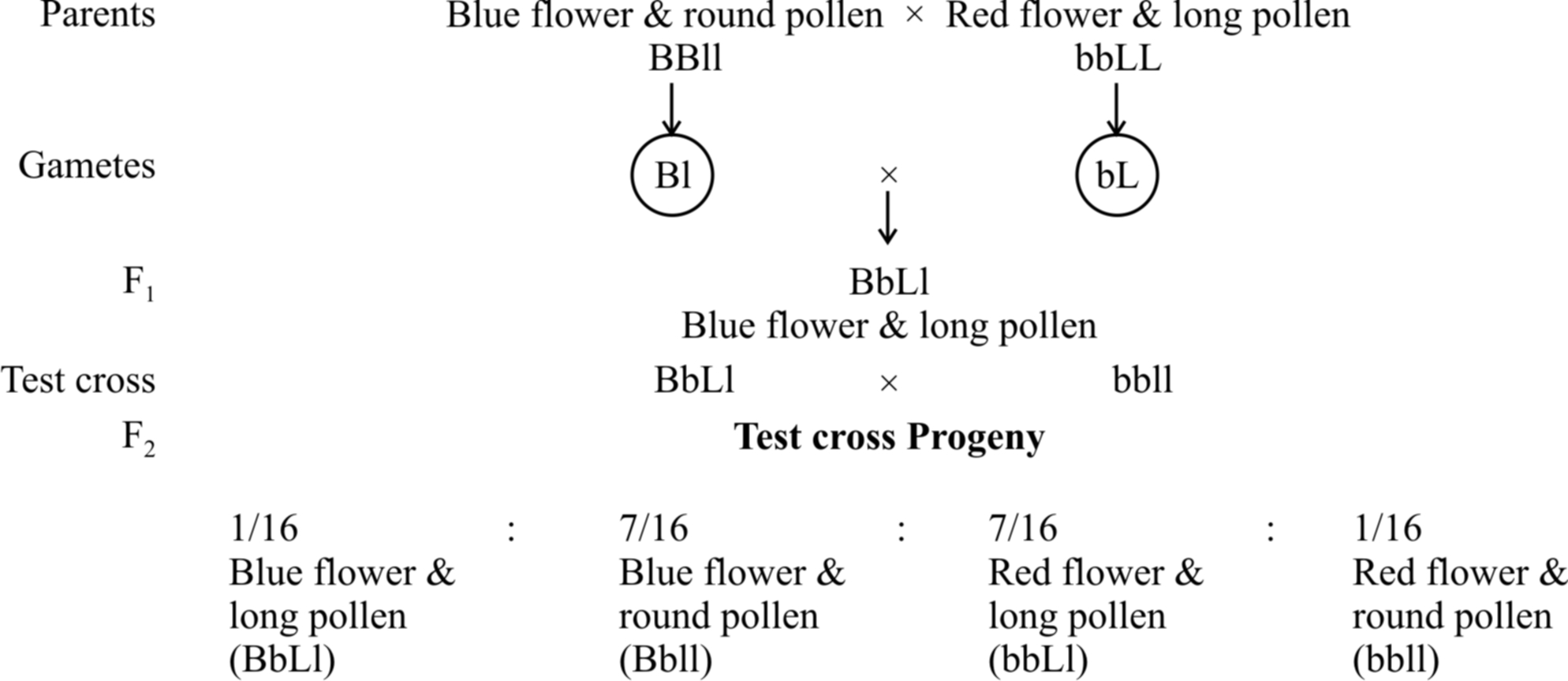
When two dominant or recessive genes come from different parents, they tend to remain separate hence, this ratio was called repulsion ratio.
T.H. Morgan in 1910 showed that coupling and repulsion are two aspects of the same phenomenon called linkage.
He suggested that the two genes present on the same chromosome, are in coupling phase and when present on two different homologous chromosomes are in repulsion phase.
Morgan carried out several dihybrid crosses in Drosophila to study genes that were sex-linked and
(A) At first, he crossed yellow bodied (y) and white eyed (w) female with brown bodied (y+) red eyed (w+) male which produced F1 with brown bodied red eyed female and yellow bodied white eyed male. In F2 generation, obtained by intercrossing of F1 hybrids, the ratio deviated significantly from expected. He found 98.7% to be parental and 1.3% as recombinants.
(B) In a second cross between white eyed and miniature winged female (wwmm) with wild red eyed (w+) normal winged male (m+), the F1 generation included red eyed normal winged female and white eyed miniature winged male. After intercrossing the F1 progeny, he found 62.8% parental and 37.2% recombinants.
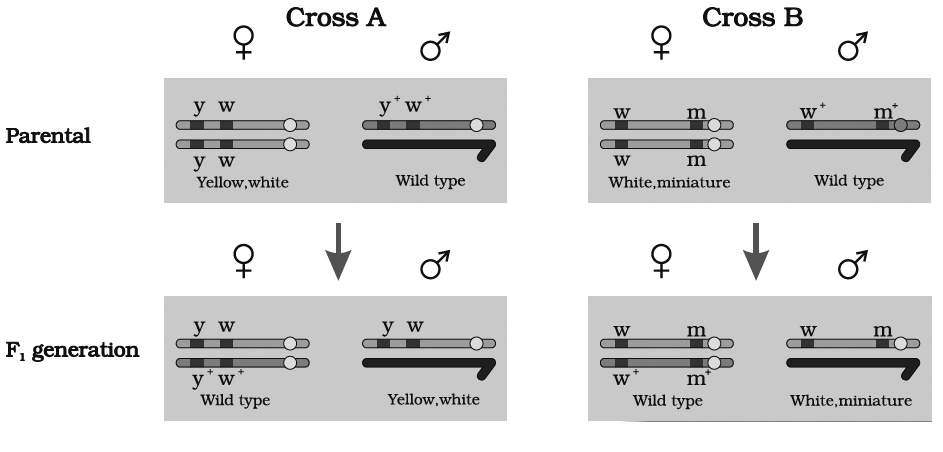
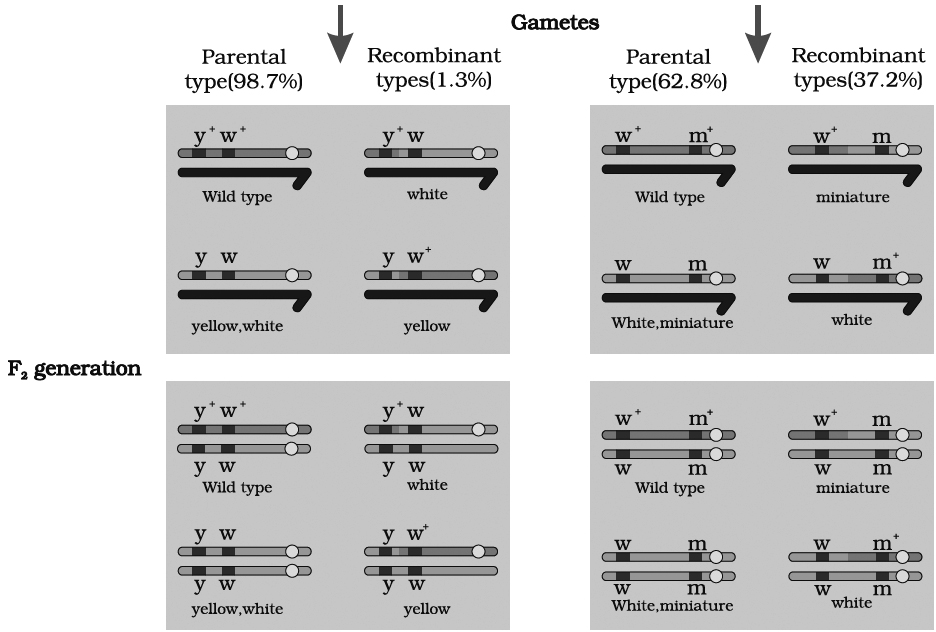
we can say that strength of linkage between y and w is higher than w and m
According to Morgan, the degree or the strength of linkage depends upon the distance between the linked genes in the chromosome.
Linkage, therefore, may be defined as "The tendency of two genes of the same chromosome to remain together in the process of inheritance".
Kinds of Linkage
T.H. Morgan and his coworkers found two types of linkage :
(1) Complete linkage.
Linkage of genes on a chromosome which is not altered and is inherited as such from generation to generation without any cross over.
In this type of linkage, genes are closely associated and tend to remain together.
Example: Male Drosophila and female silk worm (Bombax mort).
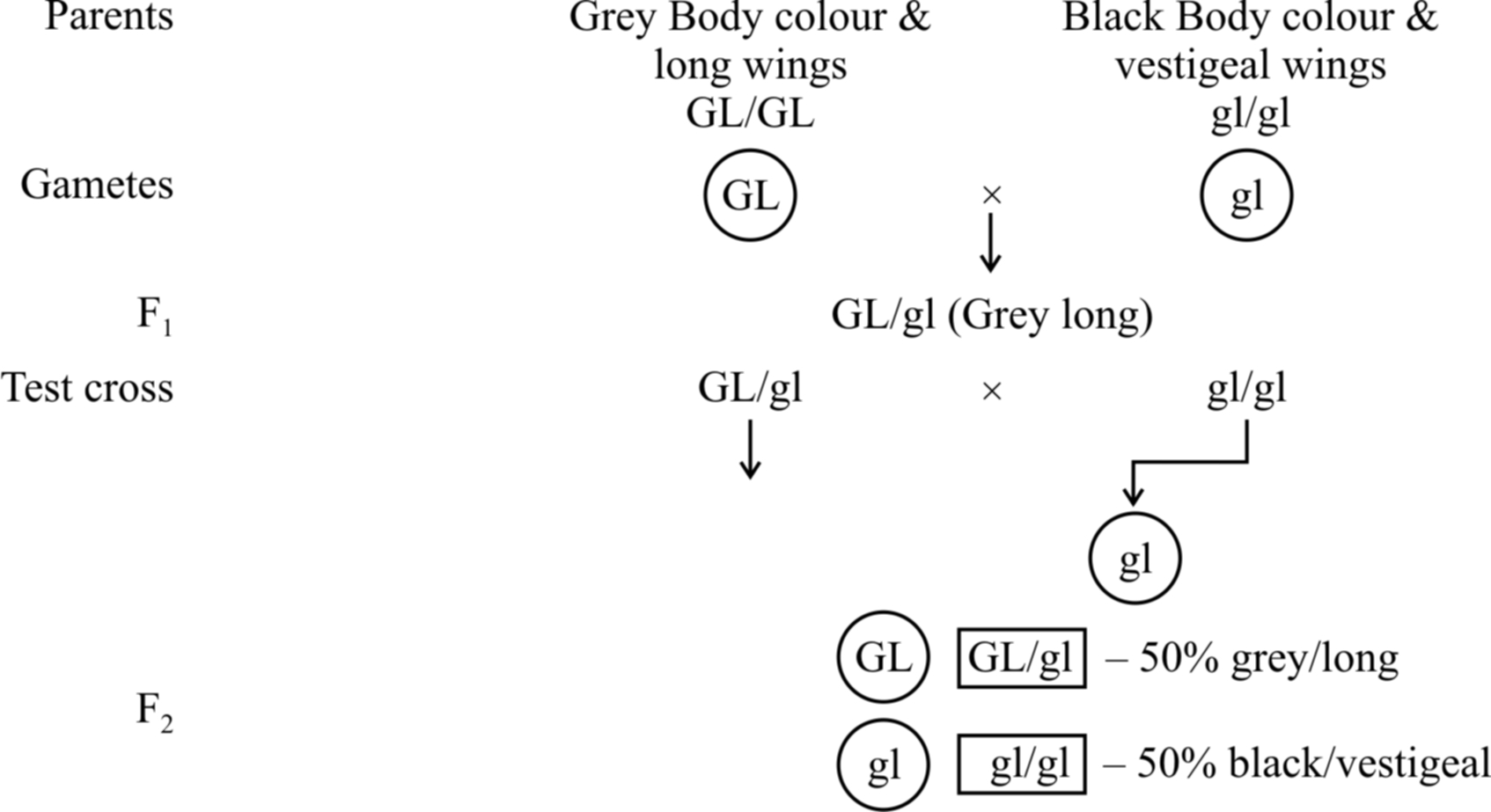
100% parental combinations indicated that the gene for grey body colour is completely linked with long wings.
In this dihybrid, F2 phenotypic ratio is 3: 1 and test cross ratio is 1 : 1 (like a monohybrid). Another example is inheritance of red eye and normal wing (PV/PV) with purple eye and vestigial wing character (pv/pv).
(2) Incomplete linkage.
The linked genes do not always stay together because homologous non sister chromatids may exchange segments of varying length with one another during meiosis.
This is known as crossing over.
The linked genes may have chances of separation by crossing over, are called incompletely linked genes and the phenomenon of their inheritance is called incomplete linkage.
It produces both parental and recombinant types in variable ratio.
Bateson and Punnet studied Lathyrus odoratus and defined coupling and repulsion of dominant and recessive gene.
In cis arrangement or coupling condition, the incomplete linkage ratio was 7 : 1 : 1 : 7 (14-parental 2-recombinants).
In trans arrangement or repulsion case, the ratio was 1:7:7:1 (parental 14, recombinants = 2).
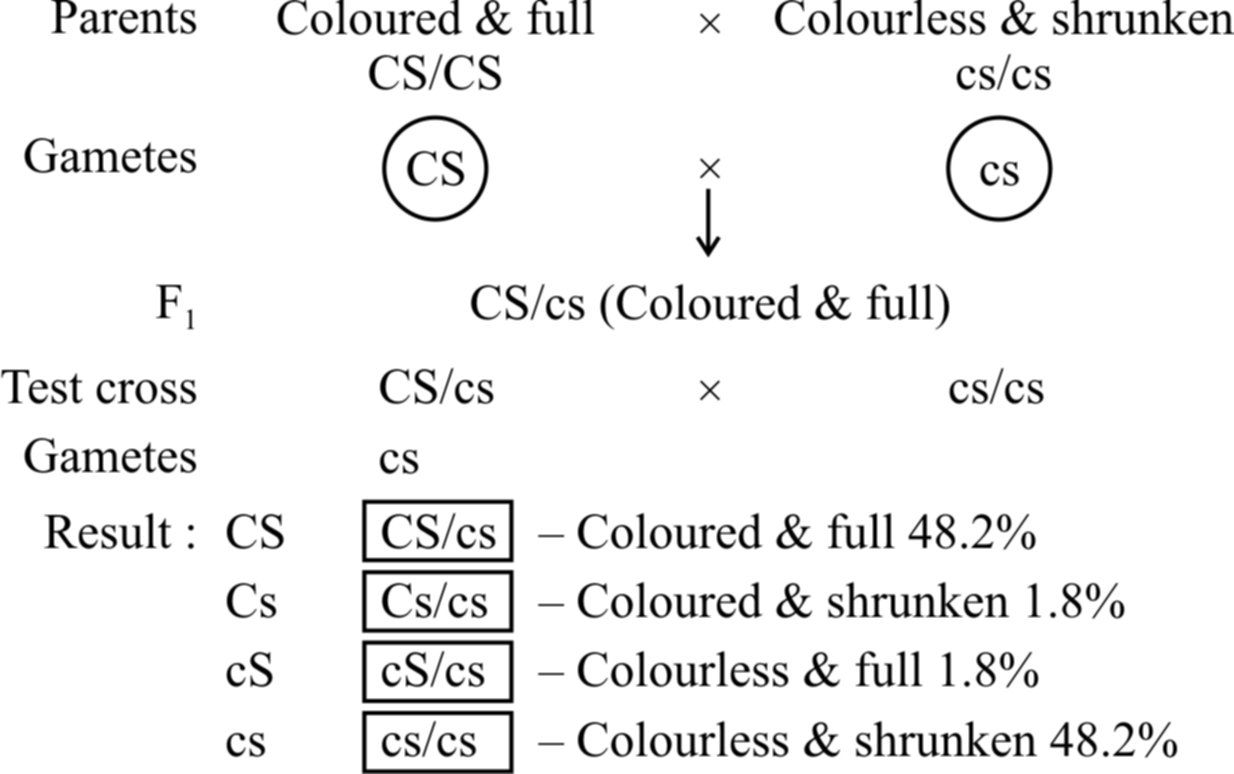
Example: In maize, incomplete linkage was observed by Hutchinson w.r.t seed coat colour and seed shape. The results show that parental combination of alleles (CS/CS and cs/cs) appear in about 96% cases. The other two are new combinations (Cs/cs and cS/cs) wh ich appear in about 4% cases. Thus, in about 4% cases, crossing over has occurred between linked genes.
Concept Builder
Linkage Group
A set of genes on a chromosome that tend to be inherited (transmitted) which can inherit together from one generation to another generation, constitute a linkage group.
Number of linkage group in an organism is equal to 'n' number of chromosome, e.g., it is 12 in rice, 7 in Pisum, while in human beings, the number of linkage group is 23 in human female and 24 in human male and in Drosophila it is 4.
CROSSING OVER AND RECOMBINATION
Crossing over is a process that produces new combination of genes by interchanging of segments between nonsister chromatids of homologous chromosomes.
The crossing over occurs in between the homologous chromosomes at four stranded or tetrad stage during pachytene of prophase I of Meiosis I.
A condition where an individual heterozygous for two pairs of linked genes (AaBb) possesses the two dominant genes on one member of the chromosome pair and the two recessive on the other, is said to be cis arrangement.

Cis arrangement
If an individual heterozygous for two pairs of linked genes (AaBb) possesses one dominant and one recessive allele of each pair of genes on each member of the homologous pair of chromosome, the arrangement is said to be Trans arrangement.

Trans arrangement
When two genes are located very close to each other in chromosomes, hardly any crossing over can be detected.
The linkage is broken down due to crossing over.
Crossing over will be relatively more frequent if the distance between two genes is more.
Frequency of crossing over can be determined cytologically by counting the number of chiasmata.
The details of the crossing over for two genes A and B and their alleles a and b on the homologous chromosomes are shown in figure.
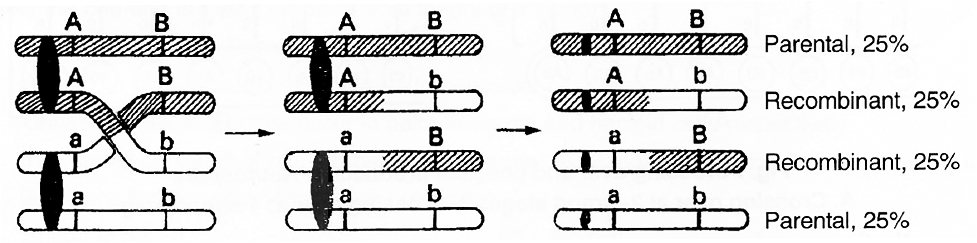
Fig. : Crossing over showing 50 per cent parental and 50 percent recombinants
The crossing over as shown above results in the formation of following four types of cells :

Crossing Over Occurs at Four Stranded Stage
Neurospora (pink mould), an ascomycetous fungus is used to demonstrate that crossing over takes place at 4-stranded stage.
It has following advantages as experimental organism :
(1) It is haploid and there is only one allele at each locus. Hence, dominant-recessive relationship does not interfere with observations and analysis.
(2) The products of single meiosis can be easily analysed.
(3) The products of meiosis occur in the form of 'ordered tetrads', i.e., the eight ascospores formed, are linearly arranged in a sac-like structure called ascus.
If genes A and B are located on same chromosome and undergo independent assortment, the genotype of linearly arranged ascospores can be studied. If crossing over takes place at 2-strand stage, the ascospores would show Ab, Ab, Ab, Ab, aB, aB, aB, aB, (4 Ab + 4 aB) arrangement.
If crossing over takes place at 4-stranded stage, the ascospores would show AB, AB, Ab, Ab, aB aB, ab, ab, (2AB + 2 Ab + 2 aB + 2 ab) arrangement or 2: 4 : 2 arrangement.
Tetrad analysis has demonstrated the presence of such an arrangement and thus, it is now confirmed that crossing over occurs at 4-stranded stage.
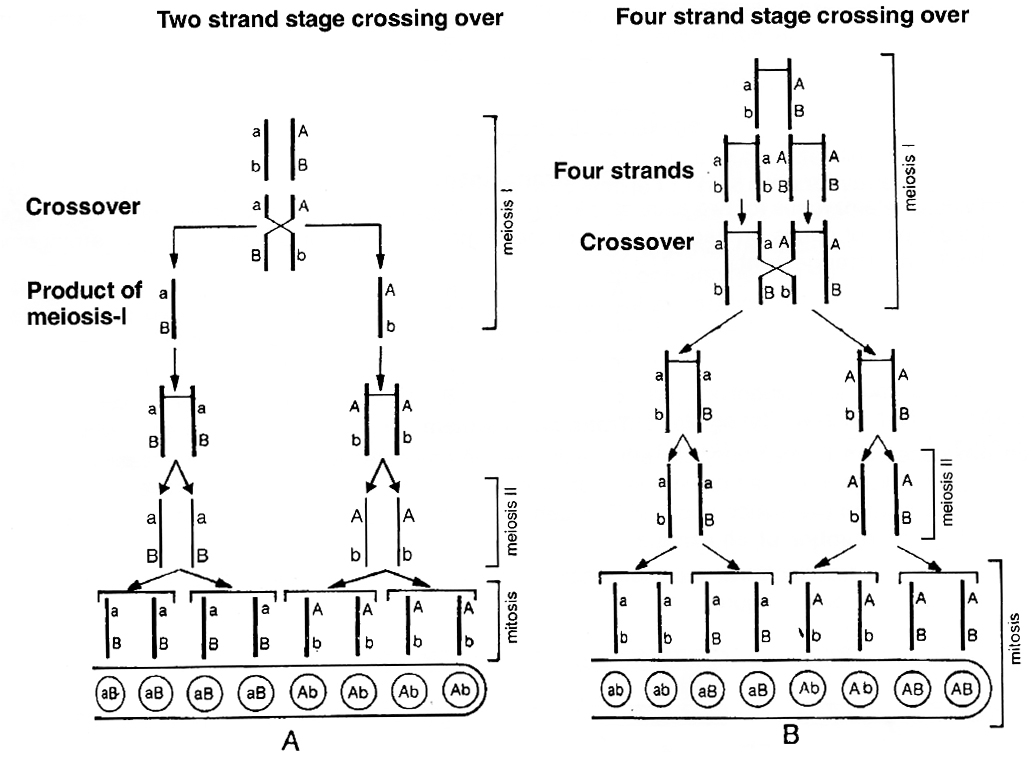
A. Crossing over at 2-strand stage; B. Crossing over at 4-strand stage
Factors affecting crossing over:
(i) Distance between the genes is directly proportional to crossing over.
(ii) Cross over decreases with age.
(iii) X-rays and temperature increases crossing over.
(iv) Centromere and heterochromatin positions decrease crossing over.
(v) One cross over reduces the frequency of other crossover in its vicinity which is called as interference.
Chromosomal Mapping
Crossing over is important in locating the genes on chromosome.
The genes are arranged linearly on the chromosome.
This sequence and the relative distances between various genes is graphically represented in terms of recombination frequencies or cross over values (COV).
This is known as linkage map of chromosome.
Distance or cross over units are called centimorgan (cM) or map unit.
Term centimorgan is used in eukaryotic genetics and map unit in prokaryotic genetics.
Recombination frequency or cross over value = 
The recombination frequency depends upon the distance between the genes.
If the distance between the genes is lesser the chances of crossing over is less and hence recombination frequency is also lesser and vice versa.
So, recombination frequency is directly proportional to the distance between genes.
In any cross, if recombination frequency is 5%.
It means the distance between the genes is 5 map unit.
A.H. Sturtevant suggested that these recombination frequencies can be utilized in predicting the sequence of genes on the chromosome.
On the basis of recombination frequency, he prepared first chromosomal map or genetic map for Drosophila.
Concept Builder
1. T.H. Morgan (1910) developed the theory of the gene as a locus or discrete unit on the chromosomes from his studies on Drosophila melanogaster.
2. Stern (1931) showed that genetic crossing over was accompanied with actual exchange of chromosome segments. He, therefore, gave the cytological proof of crossing over.
3. Map unit is related with frequency of crossing over and not with linkage.
4. C.D. Darlington -Precocity theory of crossing over.
5. J. Belling -Belling's hypothesis and copy choice mechanism of crossing over.
6. Creighton and McClintock -Cytological evidence of crossing over using corn as the material.
7. Pontecarvo (1952) -Defined genes as the ultimate unit of recombination.
8. Janssen -Discovered Chiasmata formation.
sex determination
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
SEX DETERMINATION
Sex Chromosomes and Autosomes
Sex chromosomes are those chromosomes which determine the sex of the individual in dioecious or unisexual organisms.
The normal chromosomes, other than the sex chromosomes, of an individual are known as autosomes. Sex chromosomes may be similar in one sex and dissimilar in the other.
The two conditions are respectively called homomorphic (= similar, e.g., XX, ZZ) and heteromorphic (= dissimilar, e.g., XY, ZW).
Individuals having homomorphic sex chromosomes produce only one type of gametes.
They are, therefore, called homogametic (e.g., male birds, human female and Drosophila female).
Individuals having heteromorphic sex chromosomes produce two type of gametes (e.g., X and Y).
They are termed as heterogametic (e.g., female bird, human male and normal Drosophila male).
The factors which control the sex of an organism are under genetic control.
Various mechanism which led to sex determination can be classified into following four categories:
1. Chromosomal mechanism of sex determination.
2. Non -Allosomic genetic sex determination -Fertility factor (plasmid) in bacteria.
3. Genic balance mechanism or X/A balance.
4. Environmental mechanism of sex determination.
1. Chromosomal mechanism of sex determination.
According to this mechanism, there are certain chromosomes known as sex chromosome or heterosome or idiochromosome, which are responsible for sex determination.
This may be of following types:
(i) XX -XY type:
In most insects, plants and mammals including human beings, the females possess two homomorphic (= isomorphic) sex chromosomes, i.e., XX.
The males contain two heteromorphic sex chromosomes, i.e., XY. The Y-chromosome is often shorter and heterochromatic (made of heterochromatin).
Despite differences in morphology, the Y-chromosome pairs with X-chromosome at a certain segment during meiosis.
It, therefore, carries a segment which is homologous with a segment of X-chromosome.
The remaining segment of Y-chromosome is non-homologous and carries only Y-linked or holandric genes, e.g., Testis Determining Factor (TDF).
Human beings have 22 pairs of autosomes and one pair of sex chromosomes.
All the ova (haploid) formed by female are similar in their chromosome type (22 + X). Therefore, females are homogametic.
The male gametes or sperms (haploid) produced by human males are of two types, (22 + X) and (22 + Y).
Human males are therefore, heterogametic.
The two sexes produced in the progeny may have 50 : 50 ratio.
This type of sex-determination was reported in a plant Sphaerocarpus for the first time and is also found in plants like Melandrium and Coccinia.
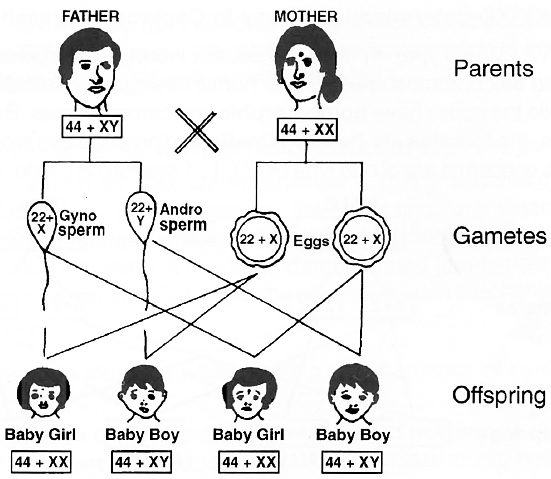
one-half of the sperms carry an X-chromosome and one-half carry a Y-chromosome.
Concept Builder
Barr Body and Y Spots
Barr and Bertram (1949) found a small darkly stained chromatin body (heterochromatic) adhered to the nuclear membrane of nerve cells of female cats but not in male cats.
Later on, Barr (1960) observed consistent presence of these chromatin bodies in epidermal cells of buccal and vaginal mucosa, skin cells etc. in females.
Lyon (1962) suggested that one X chromosome of maternal side becomes coiled, inert and hetero-pycnotic and forms a barr body.
Number of barr bodies is always one less than the total number of X chromosomes in female.
Normal male has no barr body.
In about 5% of polymorphonuclear leucocytes (neutrophils) in women, the heteropycnotic X chromosome occurs as a round pedunculated body attached to the nucleus.
It is called drum stick (Barr body).
A normal female has 1 drum stick per nucleus of neutrophil.
In neutrophils of males drum stick is absent.
Y spots: Males are identified by number of Y spots (Zech 1970). Number of Y spots are equal to number of Y Chromosomes.
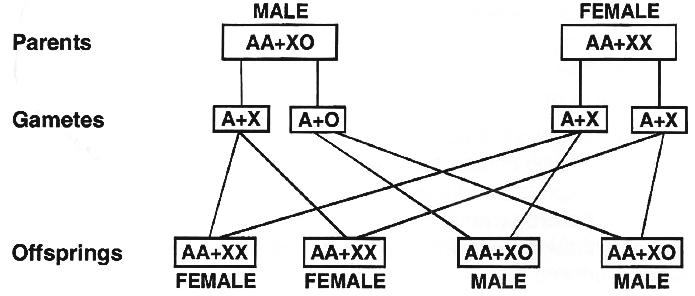
(ii) XX -XO type.
In roundworms, Dioscorea and some insects (true bugs, grasshoppers and cockroaches), the females have two sex chromosomes, XX, while the males have only one sex chromosome, X.
There is no second sex chromosome.
Therefore, the males are designated as XO.
The females are homogametic because they produce only one type of eggs.
The males are heterogametic with half the male gametes carrying X-chromosome while the other half being devoid of it.
The sex ratio produced in the progeny is 1 : 1.
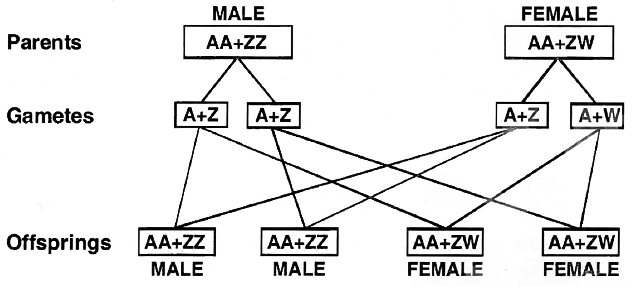
(iii) ZW -ZZ type. (WZ -WW Type).
In birds, fishes, silk worm, Fragaria elatior and some reptiles both the sexes possess two sex chromosomes.
Unlike human beings, the females contain heteromorphic sex chromosomes while the males have homomorphic sex chromosomes.
Because of having heteromorphic sex chromosomes, the females are heterogametic and produce two types of eggs, (A + Z) and (A + W).
The male gametes or sperms are of one type (A+Z). 1 : 1 sex ratio is produced in the offspring.
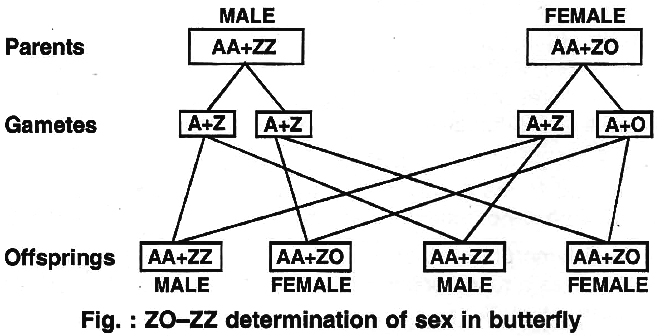
(iv) ZO -ZZ type.
This type of sex determination occurs in butterflies, pigeon, ducks and moths.
It is exactly opposite the condition found in cockroaches and grasshoppers.
Here the females have odd sex chromosome while the males have two homomorphic sex chromosomes.
The females are heterogametic.
They produce two types of eggs, with one sex chromosome (A + Z) and without the sex chromosome (A + 0).
The males are homogametic, forming similar types of sperms (A + Z).
The two sexes are obtained in the progeny in 1 : 1 ratio as both the types of eggs are produced in equal ratio.
Arrhenotoky/Haploid-diploid Mechanism:
This mechanism is found in honey bee. In honey bees, ants and wasps, the egg, if fertilized, gives rise to female fly.
The unfertilized egg develop parthenogenetically into male. So, the female flies are diploid while male flies are haploid.
2. Non-Allosomic genetic sex determination:
Fertility factor of plasmid in bacteria determines sex.
3. Genic Balance or XlA Balance Theory of Sex Determination:
Given by C.B. Bridges. According to him, Y chromosome plays no role in sex determination of Drosophila and it is the ratio between number of X-chromosome and set of autosomes which determines the sex of fly.
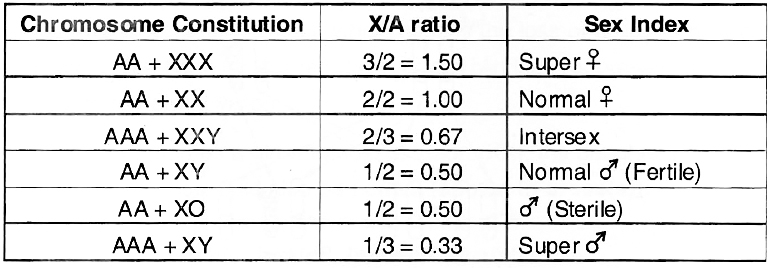
It was concluded that X/A ratio of > 1.0 expresses super femaleness, 1.0 femaleness, below 1.0 and above 0.5 intersexes, 0.5 maleness and < 0.5 supermaleness.
Gynandromorphs : Gynandromorph is a sex mosaic (an individual with one half of the body male and the other half female).
These are common in Silk moth and Drosophila. Gynandromorphism is developed due to accidental loss of X-chromosome from a 2A + XX cell during mitosis.
Gynander : A gynander may be male or female with patches of tissues of other sex on it.
4. Environmental Mechanism of Sex Determination:
This mechanism is observed by F. Baltzer in Bonnelia viridis (marine worm).
In this organism, the sex is undifferentiated in larva.
The larva which settle down in mud, grow up into mature female while those which settle down near the proboscis of female and become parasite develop into male.
It has been demonstrated that female secrete certain hormone which induces sex in larva. Crepidula and Ophryortocha also show such mechanism.
SEX LINKED INHERITANCE
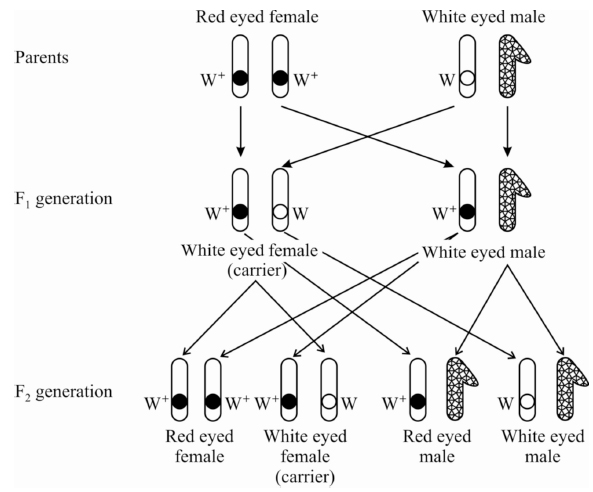
Sex linkage was discovered by Morgan, while working on inheritance of eye colour in Drosophila. He made three types of crosses:
Cross -1 :
The white eyed male (w) was crossed with red eyed (w+) female.
All the flies of F1 generation were found to be red eyed.
F1 flies were allowed to self breed.
In F2 generation, both the traits of red eye and white eye appeared in the ratio 3 : 1 showing that white eye trait is recessive to red eye trait.
Cross -2 :
Red eyed females of F1 generation were crossed with white eyed male.
It is similar to test cross where hybrids are cross bred with recessive parents.
Morgan obtained red and white eyed female as well as male in equal proportions-1 red eyed female: 1 white eyed female: 1 red eyed male: 1 white eyed male.
The test cross indicated that white eye colour was not restricted to the male fly. Red eyed White eyed hybrid female male
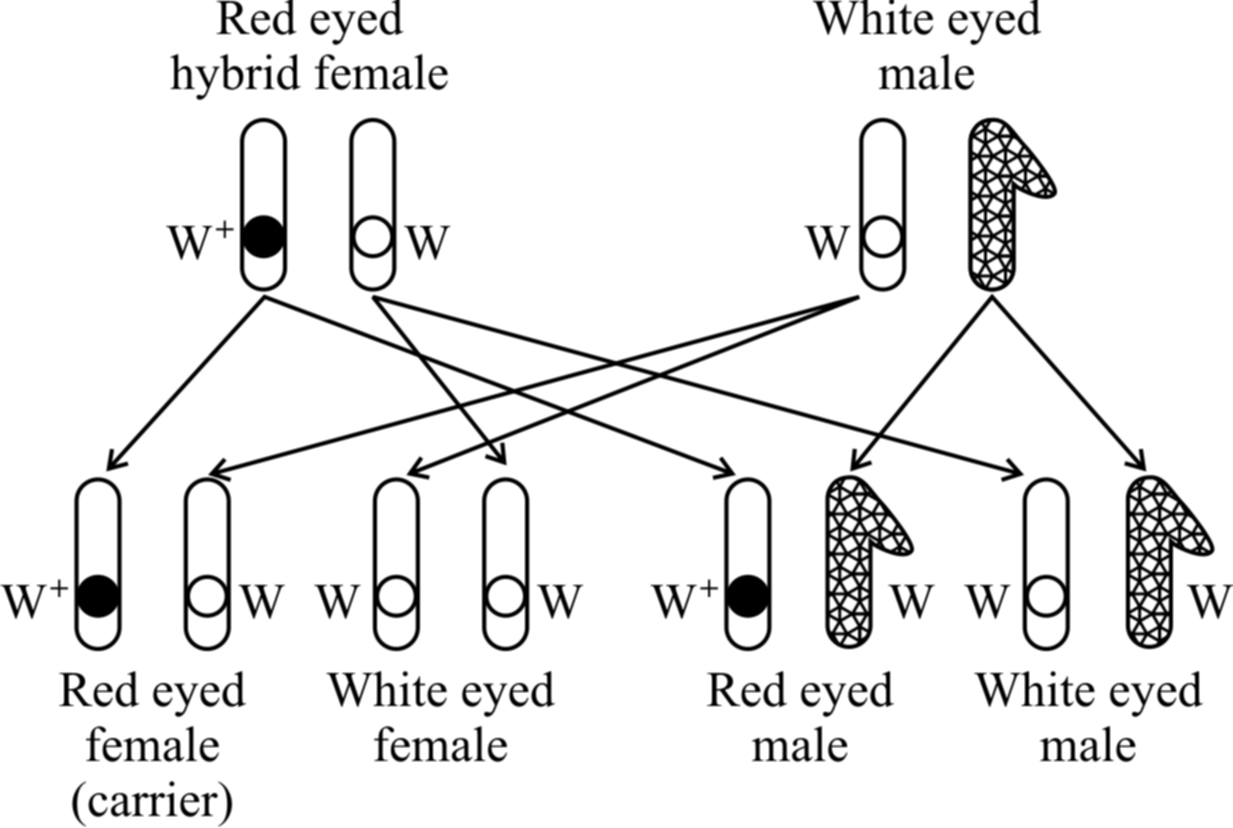
traits appear in both males and females in equal proportions
Cross -3 :
White eyed females were crossed with red eyed males. It was a reciprocal of cross 1 and should give the similar result as obtained by Mendel. However, Morgan obtained a surprising result. All the males were white eyed while all the females were red eyed.
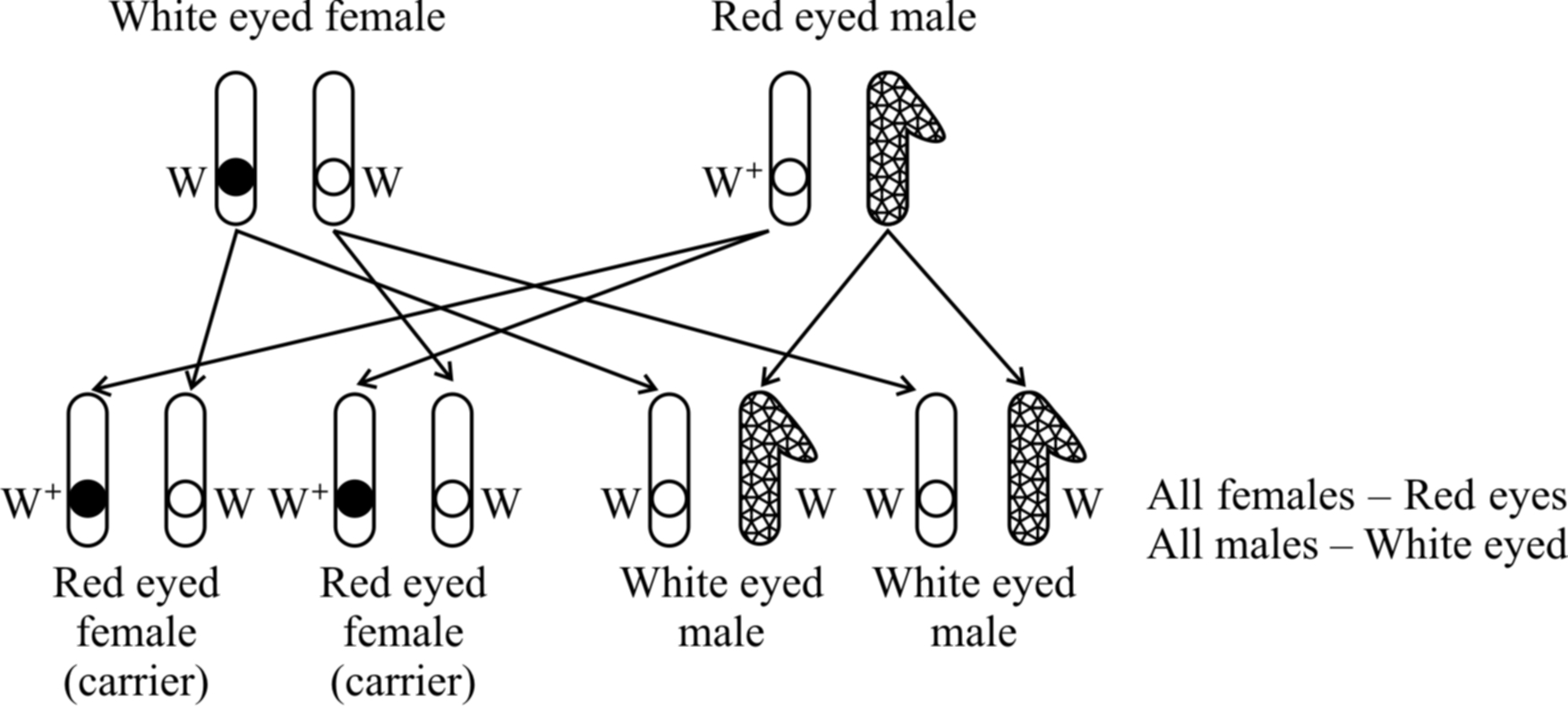
Taking all the crosses into consideration, Morgan came to the conclusion that eye colour gene is linked to sex and is present on the X-chromosome.
X-chromosome does not pass directly from one parent to the offspring of the same sex but follows a criss-cross inheritance, i.e., it is transferred from one sex to the offspring of the opposite sex.
In other words, in criss-cross inheritance a male transmits his traits to his grandson through daughter (Diagynic), while a female transmits the traits to her granddaughter through her son (Diandric).
Sex Linkage in Human Beings
Colour blindness and haemophilia (Bleeder's disease) are two common examples of sex-linked diseases in human beings.
(1) Colour blindness.
This is a human disease which causes the loss of ability to differentiate between red colour and green colour.
The gene for this red-green colour blindness is present on X chromosome. Colour blindness is recessive to normal vision.
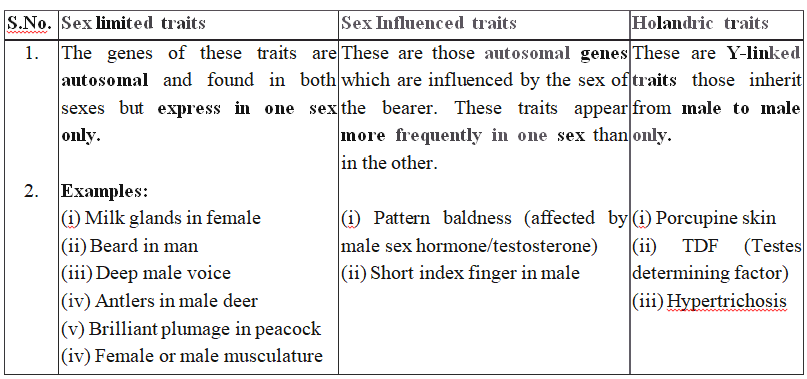
If a colour blind man (XcY) marries a girl with normal vision (XX), the daughters would have normal vision but would be carrier, while sons would also be normal (shown in cross(a)).
Cross (a)
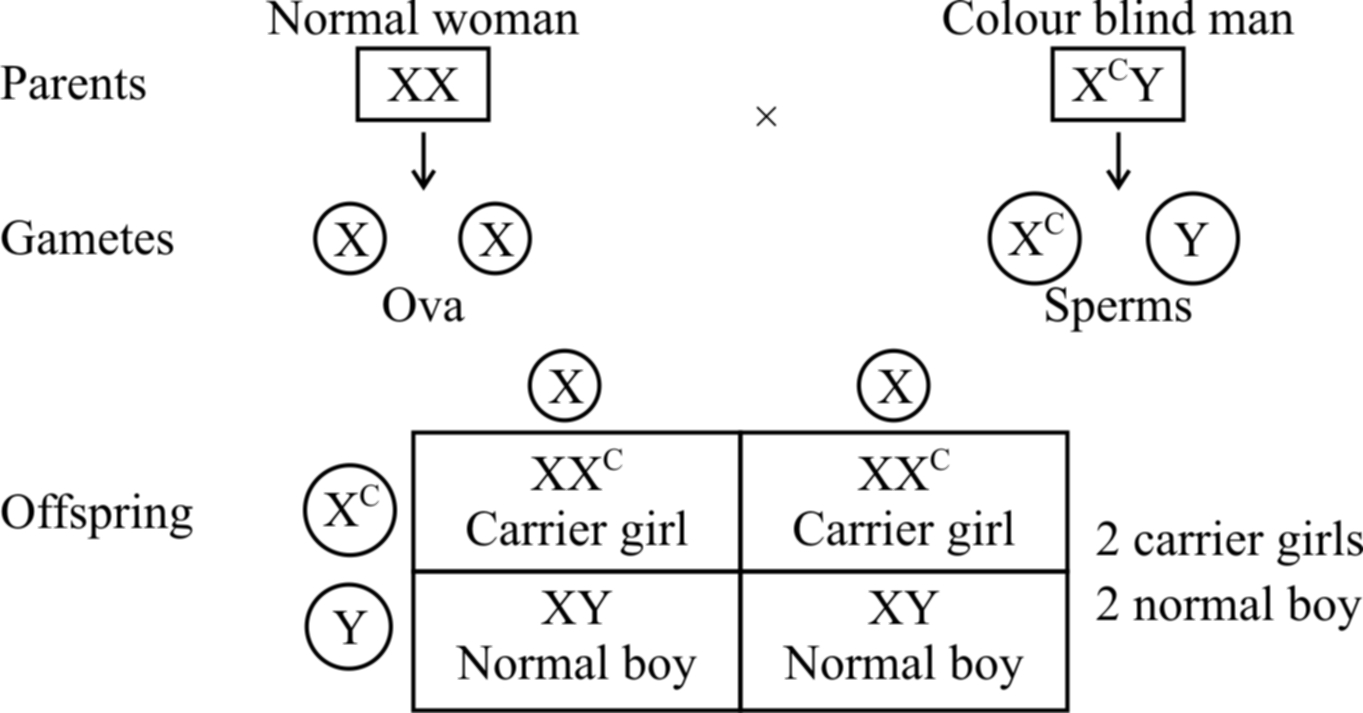
If the carrier girl (heterozygous for colour blindness, XCX) now marries a colour blind man XCY, the offspring would show 50% females and 50% males.
Of the females, 50% would be carrier for colour blindness and the rest 50% would be colour blind.
Of the males, 50% would have normal vision and the 50% would be colour blind (shown in cross (b).
Cross (b)
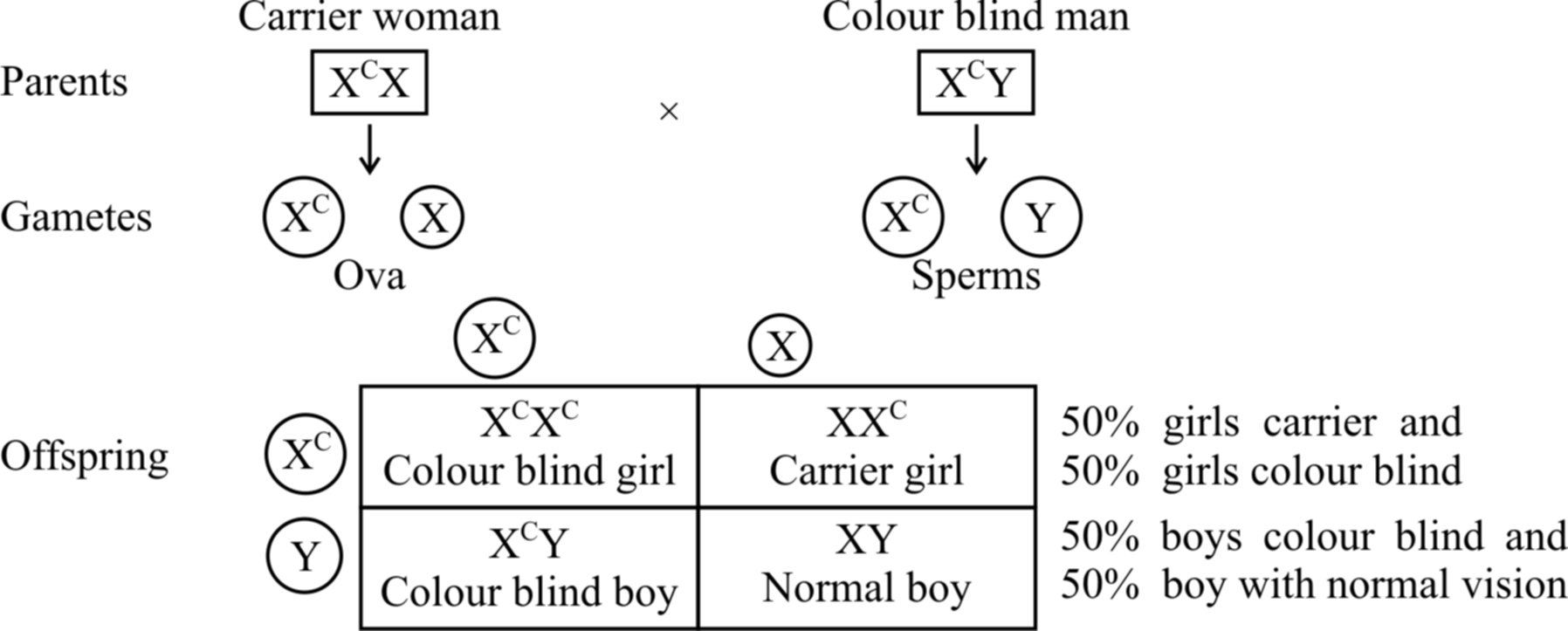
Fig. : Sex-linked inheritance of colour blindness -cross (a) and cross (b)
(2) Haemophilia (Bleeder's disease).
The person suffering from this disease cannot synthesize a normal blood protein called antihaemophilic globulin (AHG) required for normal blood clotting (Haemophilia A - more severe).
Therefore, even a very small cut may lead to continuous bleeding for a long time.
This gene is located on X chromosome and is recessive.
It remains latent in carrier females.
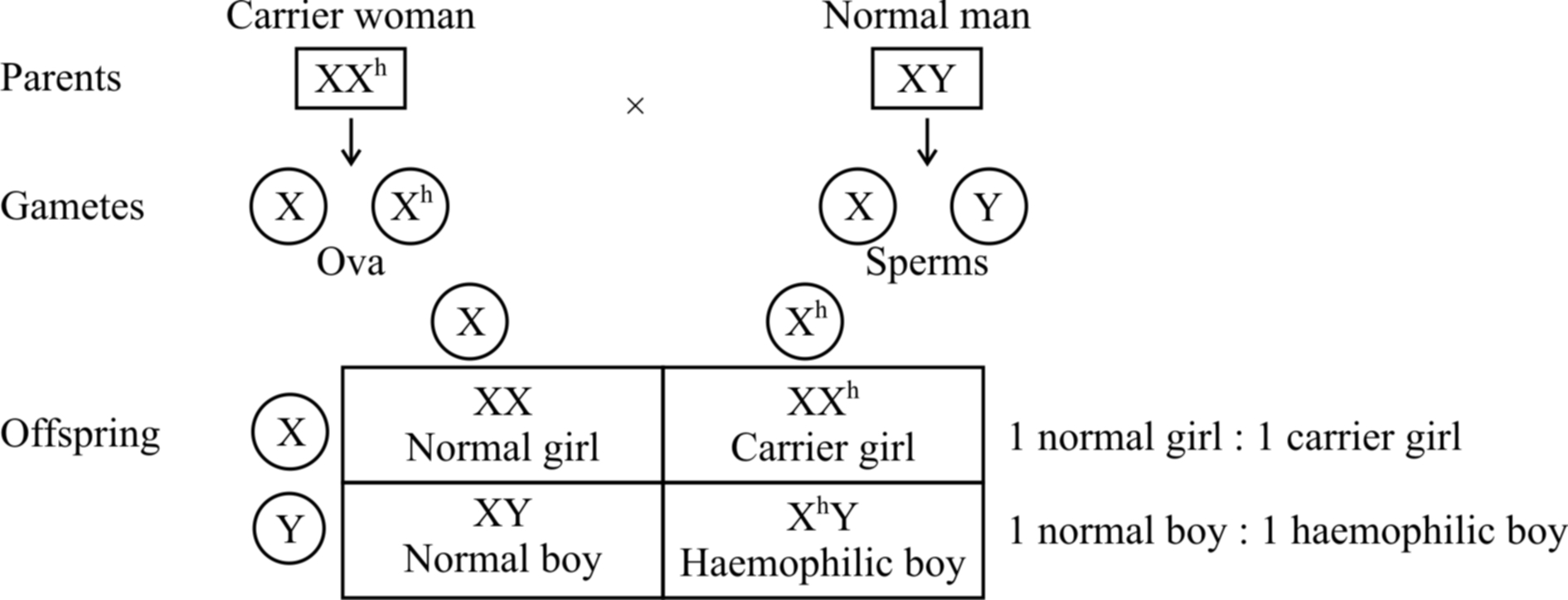
Fig.: Inheritance of haemophilia when the mother is carrier and the father is normal
If a normal man marries a girl who is carrier for haemophilia, the progeny would consist of 50% females and 50% males.
Of the females, 50% would be normal and the rest 50% would be hemophilia carrier.
Of the males, 50% would be normal and the rest would be haemophiliacs.
Haemophilia-B (christmas disease) -plasma thromboplastin is absent, Inheritance is just like Haemophilia A.
Concept Builder
1. C.E. McClung (1902) concluded that some special chromosome (X-chromosome) must be responsible for determination of sex, thus providing first definite evidence for chromosomes theory.
2. Bridges (1916) correlated cytological observations of chromosomes and inheritance of characters.
3. Y-chromosome is male determiner in man but not in Drosophila. However, its accumulation is responsible for motility of sperms in Drosophila.
4. Man is hemizygous for sex chromosomes as he has XY chromosomes.
5. Henking discovered an "X-body" in reproductive cells of firefly.
6. Y-body was discovered by Stevens.
7. Wilson and Stevens -put forward chromosome theory of sex and name X and Y body as X-chromosome and Y-chromosome.
8. Tijo and Levan -Discovered exact number of chromosomes in human beings as 46.
9. F.R. Lillie -Formation of free martins (twins in cattles).
Mutation
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
MUTATION
Mutation is sudden, inheritable, discontinuous variation due to change in chromosomes and genes.
Hugo de Vries (1901) one of the discoverers of Mendel's laws, observed two distinct varieties of Oenothera lamarckiana (Evening primrose).
These differed in length of stem, flower form and colour and shape of leaves.
These mutant varieties are now known to have been produced due to chromosomal aberrations.
Seth Wright (1791) is considered to be the first to record point or gene mutation.
He noticed a lamb with unusually short legs.
This short legged breed of sheep was known as ancon breed.
Darwin called this variation as sports.
Bateson (1894) termed them as discontinuous or saltatory variations.
The credit for starting scientific study of mutations goes to Thomas Hunt Morgan (1910).
He is known for his work on fruit fly, Drosophila melanogaster.
He found white eyed mutant of Drosophila and since then about 500 mutations have been observed by geneticists from different parts of the world.
Types of Mutations:
Different classifications of mutations are known, each based on a definite criterion or character.
I. Spontaneous and induced mutations:
These are based upon the agency involved.
Spontaneous mutations. These are natural mutations. They have also been called background mutations. Such mutations occur at a frequency of 1 × 10–5 in nature.
Induced mutations. These have been observed in organisms due to specific factors such as radiations, ultra violet light or variety of chemicals. The agents which induce mutations on their application, are called mutagens or mutagenic agents.
II. On the basis of the type of cells in which mutations occur, there are other two types of mutations.
(a) Somatic mutations. These mutations occur in somatic cells, i.e., body cells or the cells other than germinal cells. The somatic mutations do not have any genetic or evolutionary importance. This is because only the derivatives or the daughter cells formed from the mutated cell will show mutation and not the whole organism.
(b) Germinal mutations. These mutations occur in the gametes or germ cells and are also known as gametic mutations. Such mutations are heritable, and, therefore, are of great evolutionary significance. If the mutations are dominant, these are expressed in the next generation and if recessive, their phenotypic expressions remain suppressed.
III. Forward and backward mutations:
The commonest type of mutation, is the change from normal or wild type to new genotype (recessive or dominant).
Such mutations are called forward mutations.
An organism which has undergone forward mutation, may again develop mutation which restores the original wild-type phenotype.
Such reversions are known as backward mutations or reverse mutations.
Mutations can occur at any stage during the life cycle of an organism.
Types of Mutations:
1. Gene mutation.
It is alteration in the sequences of nucleotide in nucleic acids or any change in the sequence of triplet bases.
If gene mutation arise due to change in single base pair of DNA, it is called point mutation.
Gene mutation occurs by following methods :
(a) Frame-shift mutation (Gibberish mutation)
(i) Deletion : Removal of one or more bases from nucleotide chain.
(ii) Insertion or addition: Addition of one or more bases in a nucleotide chain.
(b) Substitution. The replacement of one base by another.
It is of two types :
(i) Transition: When a purine base (A or G) is substituted by another purine base or pyrimidine base (T or C) is substituted by another pyrimidine base.
(ii) Transversion: Substitution of a purine base with a pyrimidine base or vice versa.
(c) Tautomerization: The purines and pyrimidines in DNA and RNA may exist in several alternate forms or tautomers. Tautomerization occurs through rearrangement of electrons and protons in the molecule. Tautomers show changed base pairing so as to cause change in sequence like AT to CG.
Non-sense Mutation : Such mutations arise when a normal codon, coding for an amino acid is changed into a chain terminating codon (UAG, UAA, UGA) resulting in the production of an incomplete polypeptide.
Nonsense mutations rarely go unnoticed because the incomplete or shorter protein formed, is generally inactive.
Mis-sense mutation: Change in base in a codon, producing a different amino acid at the specific site in a polypeptide. In mis-sense mutation, the change in one amino acid is frequently compatible with some biological activity, e.g., sickle cell anaemia.
2. Chromosomal mutation (Chromosomal Aberrations) :
The change in the chromosome morphology is called chromosomal aberration. Structural changes in the chromosomes take place during meiosis. There are four types of chromosomal rearrangements:
(a) Deficiency :
Loss of a terminal segment of chromosome or Deletion -It is due to loss of a intercalary part of chromosome, e.g., Cri-du-Chat syndrome (short arm of chromosome 5 loses a part).
(b) Duplication:
Occurs due to addition of a part of chromosome so that a gene or set of genes is represented twice, e.g., Barr eye in Drosophila.
(c) Translocation :
It involves shifting of a part of one chromosome to another non-homologous chromosome. So new recombinant chromosomes are formed, as this induces faulty pairing of chromosomes during meiosis. An important class of translocation having evolutionary significance is known as reciprocal translocation or segmental interchanges, which involves mutual exchange of chromosome segments between non-homologous chromosome, i.e., illegitimate crossing over. Chronic myelogenous leukemia (CML) occurs due to translocation of segment of long arm from chromosome 22 to chromosome 9. Chromosome 22 is called Philadelphia chromosome.
(d) Inversion:
Change in linear order of genes by rotation of a section of chromosome by 180°. Inversion occurs frequently in Drosophila as a result of X-ray irradiation. They may be of two types:
(i) Paracentric : Inversion without involving centromere (Inverted segment does not carry centromere).
(ii) Pericentric: Inversion involving centromere.
3. Genomatic mutation or numerical changes in chromosome number :
It is of two types:
(A) Aneuploidy:
In aneuploidy, any change in number of chromosomes in an organism would be different than the multiple of basic set of chromosomes. It results due to failure of segregation of chromatids during cell division cycle. There would be following two possibilities:
(a) Hypoploidy:
This arises due to loss of one or more chromosomes or pairs of chromosome. Thus, the following conditions are likely to be produced:
(i) Monosomy (2n -1) : It is the result of loss of one chromosome for a homologous pair. Its other variant is double monosomy, i.e., (2n -1 -1).
(ii) Nullisomy (2n -2) : It is the result of loss of a complete homologous pair of chromosome.
(b) Hyperploidy: This arises due to addition of one or more chromosomes or pairs of chromosome. The following conditions are thus likely to be produced :
(i) Trisomy (2n + 1) : In this type, a single chromosome is added to the chromosome set. The trisomics were obtained for the first time in Datura stramonium (Jimson weed) by A.F. Blakeslee and his co-workers (1924). In human beings, Mongolism or Down's syndrome is due to trisomy of chromosome 21 (2n + 1 or 46 + 1, i.e., chromosome 21 is present 3 times). Others are Patau syndrome (due to trisomy of 13th) and Edward's syndrome (due to trisomy of 18th chromosome).
(ii) Tetrasomy (2n + 2) : It is the result of addition of a complete homologous pair of chromosomes (i.e., 2-chromosomes).
(B) Euploidy:
In euploidy, any change in the number of chromosomes is the multiple of the number of chromosomes in a basic set or it occurs due to variation in one or more haploid sets of chromosomes. Accordingly, these may be haploid and polyploid.
(a) Haploidy : One set of chromosomes. Haploids are better for mutation experimental studies, because all mutations either dominant or recessive can express immediately in them, as there is only one allele of each gene present in each cell.
(b) Polyploidy: Failure of cytokines is after telophase stage of cell division results in an increase in a whole set of chromosomes in an organism and this phenomenon is known as polyploidy. Polyploids are of particular importance and are, therefore, discussed below. Polyploids fall into two major categories:
(i) Autopolyploids have the same basic set of chromosomes multiplied more than twice, e.g., AAA -autotriploid, AAAA -Autotetraploid. Autopolyploids with odd number of chromosomes are seedless but show Gigantism (large size) or gigas effect (more yield and more adaptability), but are odd numbered so can only be propagated vegetatively, e.g., Banana, Pineapple. Naturally occurring autotriploids are known in banana, grapes, sugar beet, tomato and watermelons. Similarly, autotetraploids occur amongst apples, berseem, corn etc.
Autopolyploids can also be produced artificially by treating the seeds, seedlings or axillary buds with an alkaloid called colchicine. It is extracted from corm of Colchicum autumnale. The treatment produces doubling of chromosomes.
(ii) Allopolyploids are hybrids whose chromosome sets are derived from two different genomes.
Let A and B be considered as different genomes. The two diploid organisms should have AA and BB chromosome sets. The autotetraploid can now be represented as AAAA while allotetraploid is represented as AABB.
Such polyploids are the result of doubling of chromosomes in F1 hybrids derived from two different or related species.
The commonest example of allopolyploidy is Raphanobrassica developed by Russian geneticist G.O. Karpechenko (1927). A cross was made between Brassica oleracea (2n = 18) and Raphanus sativus (2n = 18). F1 hybrid produced was sterile, because chromosome sets of both these plants were dissimilar and could not pair during meiosis. However, some fertile plants with 2n = 18A + 18B (18 bivalents) were found amongst these.
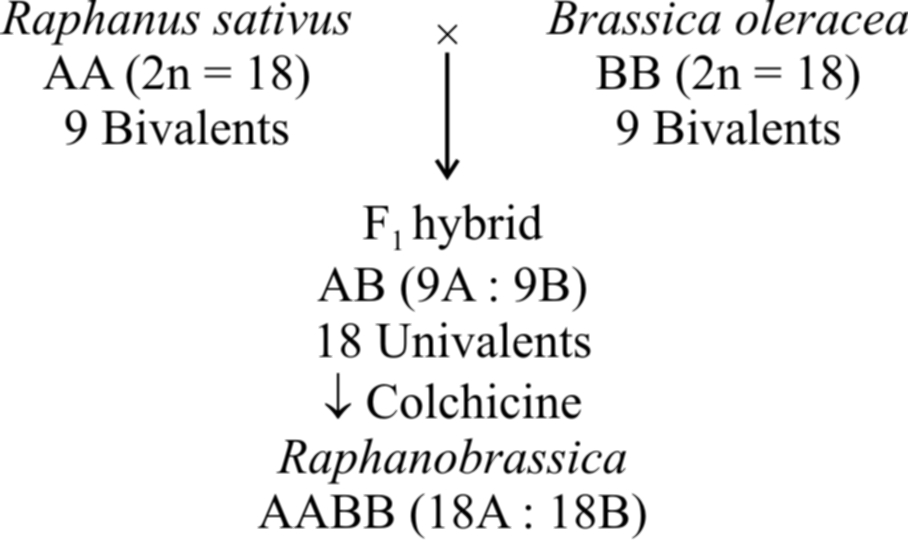
Another example is a man made cereal -Triticale, produced by (i) crossing Triticum durum (2n = 28) with Secale cereale (2n = 14) and then treating F1 hybrid with colchicine to obtain hexaploid Triticale and (ii) crossing Triticum aestivum (2n = 42) with Secale cereale (2n = 14) and then treating F 1 hybrid with colchicine to produce octaploid Triticale.
Mutagens
Mutations can be artificially produced by certain agents called mutagens or mutagenic agents. Following are two major types of mutagens :
(1) Physical mutagens
(2) Chemical mutagens
(1) Physical mutagens:
Radiations are the most important physical mutagens.
H.J. Muller who used X-rays, for the first time, to increase the rate of mutation in Drosophila, opened an entirely new field in inducing mutations.
So, Muller is considered as father of Actinobiology.
The main source of spontaneous mutations are the natural radiations coming from cosmic rays of the sun.
The spectrum of wavelengths that are shorter (i.e., of higher energy) than the visible light can be subdivided into following two groups.
(a) Ionizing radiations
(b) Non-ionizing radiations
They occur in small amounts in the environment and are known as background radiations.
The following are biological effects of radiations :
(a) Effects of ionizing radiations: The ionizing radiations include X-rays, -rays, -rays and -rays. Ionizing radiations cause breaks in the chromosome. These cells then show abnormal cell divisions. If these include gametes, they may be abnormal and even die prematurely. Different types of cancers may result due to radiations. The frequency of induced mutations is directly proportional to the doses of radiations.
(b) Effects of non-ionising radiations : The non-ionizing radiations have longer wavelengths but carry lower energy. This energy is insufficient to induce ionization. Therefore, non-ionizing radiations such as UV light do not penetrate beyond the human skin . Thymine (pyrimidine) dimer formation is a major mutagenic effect of UV rays that disturbs DNA double helix and thus, DNA replication.
(2) Chemical mutagens:
Large number of chemical mutagens are now known. These are more injurious than radiations. The first chemical mutagen used was mustard gas by C. Auerbach et. al. during world war II.
Chemical mutagens are placed into two groups:
(a) Those which are mutagenic to both replicating and non-replicating DNA such as nitrous acid,
(b) Those which are mutagenic only to replicating DNA, such as acridine dyes and base analogues.
Following are the effects of some of the chemical mutagens:
(i) Nitrous acid: It is mutagenic to both replicating and non-replicating DNA. It acts directly by oxidative deamination on A, G and C bases which contain amino groups. A is deaminated to hypoxanthine which is complementary to cytosine. G is converted to xanthine which pairs with C. Cytosine is converted to U which pairs with A
(ii) Acridines: Acridines and proflavins are very powerful mutagens. These can intercalate between DNA bases and interfere with DNA replication, producing insertion or deletion or both of single bases respectively. This induces frame shift-mutations or gibberish mutation, e.g., Thalassaemia.
(iii) Base analogues: These have structures similar to the normal bases and are incorporated into DNA only during DNA replication. Base analogues cause mispairing and eventually give rise to mutations. The base analogues may either be natural or artificial. Natural base analogues include 5-methyl cytosine, 5-hydroxymethyl cytosine, 6-methyl purine etc.
The most commonly used artificial base analogues are 5-bromouracil and 2-aminopurine. 5-bromouracil is a structural analogue of thymine. It gets incorporated into the newly replicated DNA in place of thymine (T). 2-aminopurine is an artificial base analogue of adenine. It acts as a substitute of adenine (A) and can also pair with cytosine (C).
CYTOPLASMIC INHERITANCE
Some self replicating genes (DNA) are present in the cytoplasm (mitochondrial DNA and chloroplast DNA) also.
These are called plasmagenes and all the plasmagenes together constitute plasmon (like genome).
The inheritance of characters by plasmagenes is called extranuclear or extrachromosomal inheritance.
Certain most important examples of extranuclear inheritance in eukaryotes are following :
Maternal inheritance:
The amount of nuclear hereditary material contributed by the two sexes is almost equal but the cytoplasm in egg is always much more than that of the sperm.
So, in extranuclear inheritance, contribution of female parent is more.
This is called maternal inheritance.
The evidence of maternal inheritance is the coiling of shell in snails.
Concept Builder
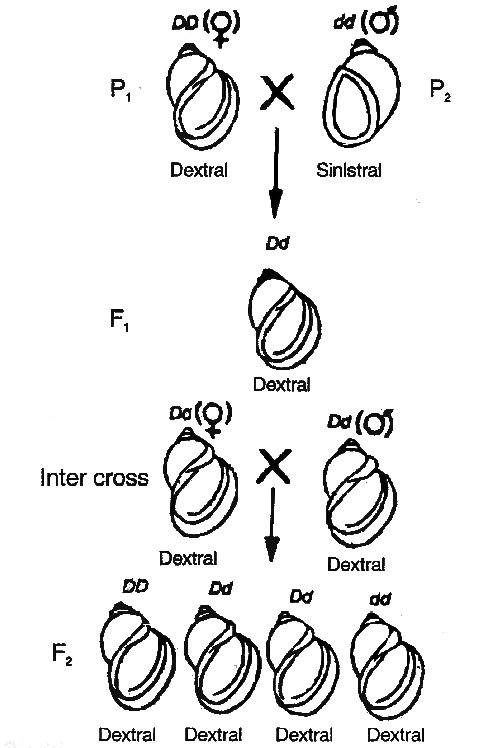
Shell Coiling in Snails
Shell coiling in Limnaea peregra, a fresh water snail, is of two types, dextral ( =clockwise) and sinistral (anticlockwise).
 dextral (DD) and
dextral (DD) and  sinistral (dd) snails
sinistral (dd) snailsDextral coiling is controlled by a dominant nuclear gene called D while sinistral coiling is due to a recessive gene d.
Since, zygote receives whole of its cytoplasm from the egg, the direction of shell coiling in the offspring is governed by cytoplasm of the mother.
When dextral female (DD) is crossed with sinistral male (dd), all the offsprings of F1 generation (Dd) have dextral coiling.
If sinistral female (dd) is crossed with dextral male (DD), the offspring have Dd genotype but coiling is sinistral.
When the latter are self-bred, three types of genotypes are produced1DD, 2Dd and 1dd.
However, shell coiling in all is dextral because the mother has Dd genotype.
Organelle inheritance:
The DNA is present in mitochondria and chloroplast which controls the inheritance of some characters.
A well known example of the characters controlled by chloroplasts is plastid inheritance in Mirabilis jalapa (4 O'clock plant), discovered by Correns.
Other examples of organellar inheritance are iojap inheritance in maize, inheritance of poky (imbalance in the mitochondrial physiology) in the fungus Neurospora crassa and Petite in yeast, a mitochondrial character.
Cytoplasmic male sterility in maize, is also a function of defective mitochondria.
Genetic disorders
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
GENETIC DISORDERS
Pedigree Analysis :
A record of inheritance of certain genetic traits for two or more generations presented in the form of a diagram or family tree is called pedigree.
Parents are shown by horizontal line while their offsprings are connected to it by a vertical line.
The offsprings are also shown in the form of a horizontal line below the parents and numbered with arabic numerals.
Pedigree analysis is study of pedigree for the transmission of particular trait and finding the possibility of absence or presence of that trait in homozygous or heterozygous state in a particular individual.
It is useful for the genetic counsellors to advise intending couples about the possibility of having children with genetic defects like haemophilia, colour blindness, alkaptonuria, phenylketonuria, thalassaemia, sickle cell anaemia (recessive traits), brachydactyly, myotonic dystrophy and polydactyly (dominant traits).
Pedigree analysis indicates that Mendel's principles are also applicable to human genetics with some modifications found out later, like quantitative inheritance, sex linked characters and other linkages.
Symbols used in Pedigree analysis :
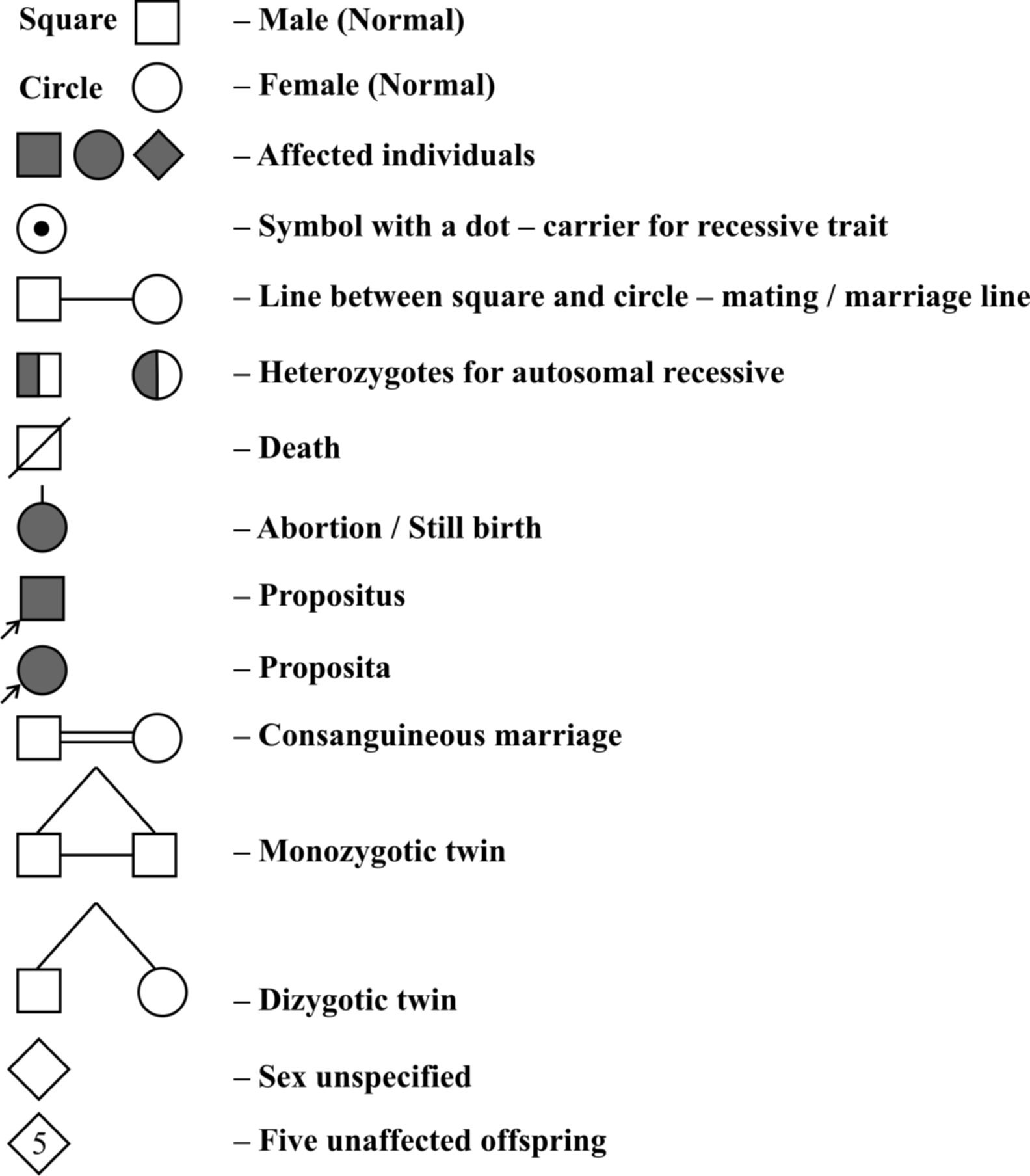
Proband is person from which case history starts. If it is male, it is called propositus, if it is female it is called proposita.
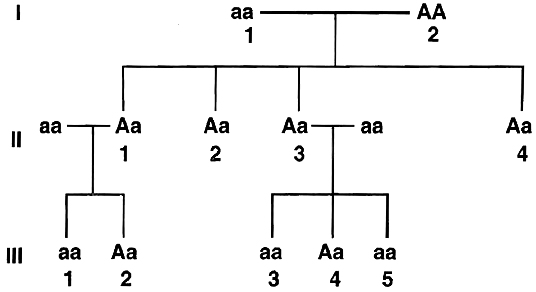
Example 8 : In the pedigree given below, indicate whether the shaded symbols indicate dominant or recessive allele. Also give genotype of the whole pedigree.
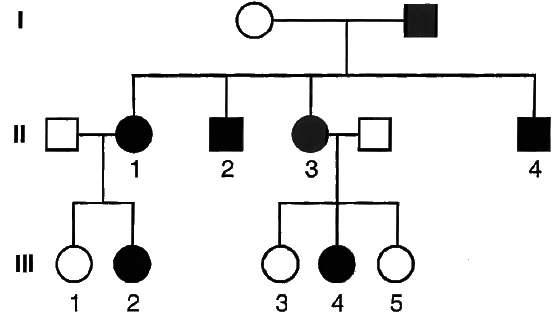
Solution: Since the shaded symbol appears in all the offsprings, father must be homozygous dominant while the mother homozygous recessive (AA × aa = all Aa) because in all other cases this possibility is absent (Aa × aa = 2Aa + 2aa; aa × AA = all Aa; aa × Aa = 2aA + 2aa). All the members of II generation will, therefore, be heterozygous (Aa). This is further confirmed by marriage of II-1 with homozygous recessive (Aa × aa = 2Aa, 2aa) and bearing children of both the parental types. Marriage of II-3 with the homozygous recessive can produce both recessive and heterozygotes as are present here.
MENDELIAN DISORDERS
Sickle-Cell Anaemia
It is an autosomal recessive disorder. In this disorder, the RBCs become sickle shaped under low O2 concentration.
The affected persons die young.
Other heterozygous for this trait are having normal phenotype and long lived.
The disease is due to base substitution of sixth codon in the gene coding for chain of haemoglobin.
The middle base of a DNA triplet coding for the amino acid glutamic acid is mutated so that the triplet now codes for valine instead.
The mutant haemoglobin molecule undergoes polymerisation under low O2 tension causing the change in the shape of RBC from biconcave disc to elongated sickle like structure.
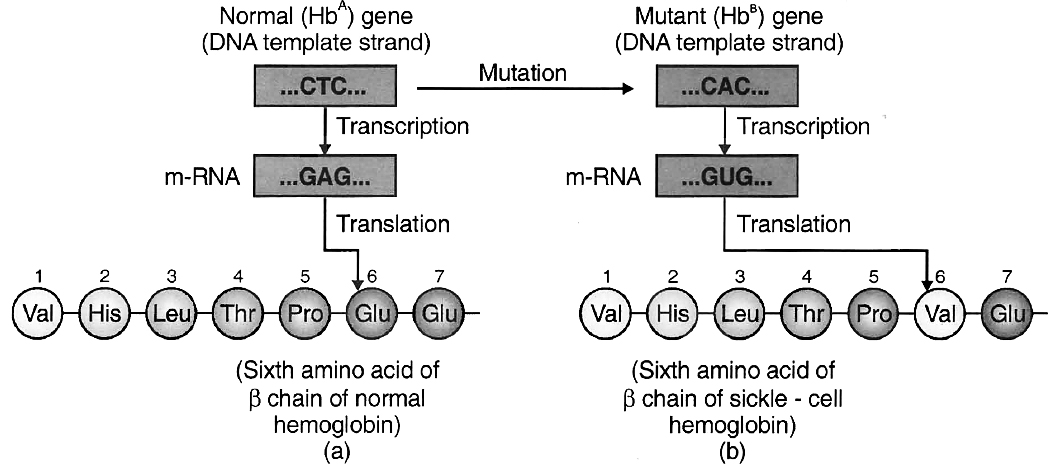
(a) from a normal individual; (b) from an individual with sickle-cell anaemia.
Thalassaemia
It is recessive autosomal disease caused due to reduced synthesis of or polypeptide of haemoglobin. thalassaemia is a major problem, individuals suffering from major thalassaemia often die before ten years of age.
Phenylketonuria
Recessive autosomal disorder (Chromosome 12) related to phenylalanine metabolism to tyrosine. This disorder is due to absence of a liver enzyme called phenylalanine hydroxylase. Due to lack of this enzyme, phenylalanine follows another pathway and gets converted into phenylpyruvic acid. This phenyl pyruvic acid upon accumulation in joints causes arthritis and if it hits the brain, then it causes mental retardation known as phenyl pyruvic idiocy. These are also excreted through urine because of poor absorption by kidney.
Cystic Fibrosis
It is an autosomal recessive disorder common among Caucasian Northern Europeans. Persons suffering from this disease are having extremely salty sweat. It is due to decreased Na+ and Cl– reabsorption in the ducts. Disease is due to a gene present on chromosome 7. Due to a defective glycoprotein, thick mucus develops in pancreas and lungs and formation of fibrous cyst occurs in pancreas.
Huntington's Chorea
It is an autosomal dominant disorder. The gene responsible for this disorder is present on chromosome 4. Disease is characterised by gradual degradation of brain tissue in the middle age and consequent shrinkage of brain.
Alzheimer's Disease
This autosomal recessive disease results in mental deterioration (loss of memory, confusion, anxiety) and ultimately the loss of functional capacities. The disease is due to deposits of -amyloid, a short protein in brain which results in degradation of neurons. It involves two defective alleles located on chromosome number 19 and 21. This disease is common in Down's syndrome.
Myotonic dystrophy is due to a dominant autosomal mutant gene located on the long arm of chromosome 19. Mild myotonia -atrophy and weakness of the musculature of the face and extremities, is most common.
Other Mendelian disorders :
(i) Alkaptonuria (Garrod, 1908) - Due to deficiency of oxidase enzyme.
(ii) Albinism (Chromosome 11) - Absence of tyrosinase
(iii) Tay-Sach's disease (Chromosome 15) - Absence of hexosaminidase B.
(iv) Gaucher's disease (Chromosomes 1) - Due to the inhibition of glucocerebrosidase enzyme action which leads to accumulation of cerebroside.
Other abnormalities due to autosomal dominant gene mutation
(i) Polydactyly - Presence of extra fingers and toes
(ii) Brachydactyly - Abnormal short fingers and toes
Abnormalities due to sex linked (X-linked) recessive gene mutation
(i) Haemophilia A - Due to lack of antihaemophilic-globulin.
Haemophilia B - Due to lack of plasma thromboplastin
(ii) Red-green colour blindness - Daltonism
Protanopia - Red colour blindness
Tritanopia - Blue colour blindness
Deuteranopia - Green colour blindness
(iii) Muscular dystrophy - Due to non-synthesis of protein dystrophin
Deterioration of muscles at an early stage
(iv) Lesch Nyhan syndrome -Deterioration of nervous system
Due to HGPRT deficiency (Hypoxanthin guanine
phosphoribosyl transferase)
CHROMOSOMAL DISORDERS
A. Autosomal abnormalities (Due to mutation in body chromosome)
(i) Down's Syndrome -It occurs due to trisomy of 21st chromosome. The affected individual is short statured with small round head, furrowed tongue and partially open mouth. Palm is broad with characteristic palm crease and mental retardation. Physical and psychomotor development is retarded.
(ii) Edward's syndrome - Trisomy of 18th chromosome
(iii) Patau's syndrome - Trisomy of 13th chromosome
(iv) Cri du chat syndrome - Due to deletion in short arm of 5th chromosome.
B. Allosomal or Sex Chromosomal Disorder
(i) Klinefelter's Syndrome - It occurs due to the trisomy of X-chromosome in male, resulting into a karyotype of 47, (44 + XXY). Individuals have long legs, sparse body hair, small prostrate gland, small testes, reduced mental intelligence and enlarged breasts (Gynaecomastia). Such individuals are sterile.
(ii) Turner's Syndrome - It is caused due to absence of one of the X-chromosome in female i.e. 45 with chromosome complement 44 + XO. Such females are sterile with undeveloped breast, short stature, reduced ovaries & absence of menstrual cycle.
(iii) Super female -AA + XXX, AA + XXXX
(iv) Jacob's syndrome or Super male -AA + XYY, also called as criminal syndrome.
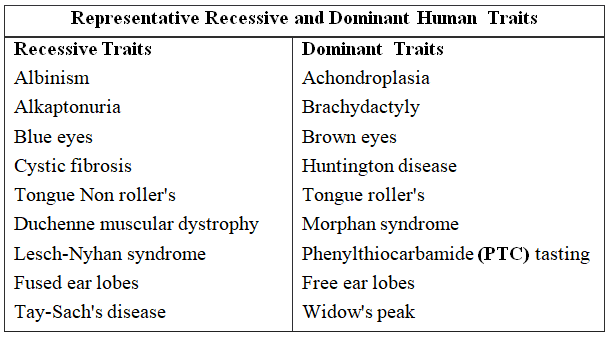
Population Genetics
Hardy Weinberg equation is applied to know the distribution of traits and frequency of autosomal dominant recessive gene distribution in the entire population.
p = Dominant gene/allele
q = Recessive gene/allele
p + q = 1
(p + q)2 = p2 + q2 + 2pq = 1
 (p2 + 2pq) = Frequency of dominant trait.
(p2 + 2pq) = Frequency of dominant trait.
 (q2) = Frequency of recessive trait.
(q2) = Frequency of recessive trait.
Concept Builder
1. Archibald Garrod discovered "Inborn errors of metabolism in humans" -phenylketonuria, alkaptonuria and albinism.
2. Muscular dystrophy and haemophilia causing genes are sex-linked recessive lethal genes.
3. The symptomatic treatment of genetic diseases of man is called Euphenics.
4. Improvement of human race by improving environmental conditions is called Euthenics.
5. Improvement of the future qualities of mankind by selective breeding is called Eugenics.
6. F. Galton -Term "Eugenics".
7. Lejeune -Discovered Cri-du-chat syndrome
8. Darligton (1939) -The genes present in the cytoplasm are called plasmagenes.
structure of DNA
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
The DNA
DNA is a long polymer of deoxyribonucleotides.
The length of DNA is usually defined as number of nucleotides or a pair of nucelotide referred to as base pairs present in it.
This also is the characteristic of an organism.
Few examples are sited below :

Chemical structure of DNA polynucleotide :
The basic unit of DNA is a nucleotide which has three components -a nitrogenous base, a pentose sugar (deoxyribose), and a phosphate group.Chemical structure of DNA polynucleotide :
There are two types of nitrogenous bases - Purines (Adenine and Guanine), and Pyrimidines (Cytosine and Thymine).
Cytosine is common for both DNA and RNA and Thymine is present in DNA.
Uracil is present in RNA at the place of Thymine.
A nitrogenous base is linked to the pentose sugar through a N-glycosidic linkage to form a nucleoside, such as adenosine or deoxyadenosine, guanosine or deoxyguanosine, cytidine or deoxycytidine and deoxythymidine.
When a phosphate group is linked to 5'-OH of a nucleoside through phosphoester linkage, a corresponding nucleotide (or deoxynucleotide depending upon the type of sugar present) is formed.
Two nucleotides are linked through 3'-5' phosphodiester linkage to form a dinucleotide.
More nucleotides can be joined in such a manner to form a polynucleotide chain.
A polymer thus formed, has a free phosphate moiety at 5'-end of sugar, which is referred to as 5'-end of polynucleotide chain.
Similarly, at the other end of the polymer the sugar has a free 3'-OH group which is referred to as 3'-end of the polynucleotide chain.
The backbone in a polynucleotide chain is formed due to sugar and phosphates (phosphodiester bond) nitrogenous bases linked to sugar moiet projects from the backbone.
In RNA, every nucleotide residue has an additional -OH group present at 2' -position in the ribose.
Also, in RNA the uracil is found at the place of thymine (S-methyl uracil, another chemical name for thymine).
In 1953, James Watson and Francis Crick based on the X-ray diffraction data produced by Maurice Wilkins and Rosalind Franklin, proposed Double Helix model for the structure of DNA.
One of the hallmarks of their proposition was base pairing between the two strands of polynucleotide chain.
However, this proposition was also based on the observation of Erwin Chargaff –that for a double stranded DNA, the ratios between Adenine and Thymine and Guanine and Cytosine are constant and equal.
Chargaff (1950) made observations on the bases and other contents of DNA. These observations or generalizations are called Chargaff's rules.
(i) Purine and pyrimidine base pairs are in equal amount, i.e., adenine + guanine = thymine + cytosine.
(ii) Molar amount of purine (adenine) is always equal to the molar amount of pyrimidine (thymine). Similarly, guanine is equalled by cytosine.
(iii) Sugar deoxyribose and phosphate occur in equimolar proportions.
(iv) The ratio of A + T/G + C is constant for a species (Base ratio, e.g., 1.52 for human and 0.93 for E.coli.
The base pairing is a very unique property of the polynucleotide chains.
They are said to be complementary to each other, and therefore if the sequence of bases in one strand is known then the sequence in other strand can be predicted.
Thus, if one DNA strand has A, the other would have T and if one has G, the other, would have C.
Therefore, if the base sequence of one strand is CAT TAG GAC, the base sequence of other strand would be GTA ATC CTG.
Hence, the two polynucleotide strands are called complementary to one another.
Also, if each strand from a DNA or parental DNA acts as a template for synthesis of a new strand, the two double stranded DNA or daughter DNA produced would be identical to the parental DNA molecule.
Salient features of DNA Double Helix
(i) It is made of two polynucleotide chains, where the backbone is constituted by sugar-phosphate, and the bases project inside.
(ii) The two chains have anti-parallel polarity. It means, if one chain has the polarity 5P 3OH, the other has 3OH 5P.
(iii) The bases in two strands are paired through hydrogen bond (H-bonds), forming base pairs(bp). Adenine forms two hydrogen bonds with Thymine from opposite strand and vice-versa. Guanine is bonded with Cytosine with three H-bonds. As a result, always a purine comes opposite to a pyrimidine. This generates approximately uniform distance between the two strands of the helix.
(iv) The plane of one base pair stacks over the other in double helix. This, in addition to H-bonds, confers stability to the helical structure.
(v) The two chains are coiled in a right-handed fashion. The pitch of the helix is 3.4 nm and there are roughly 10 bp in each turn. Consequently, the distance between base pairs in a helix is approximately equal to 0.34 nm. It is because of specific base pairing with a purine lying opposite to pyrimidine which makes two chains 2 nm thick.
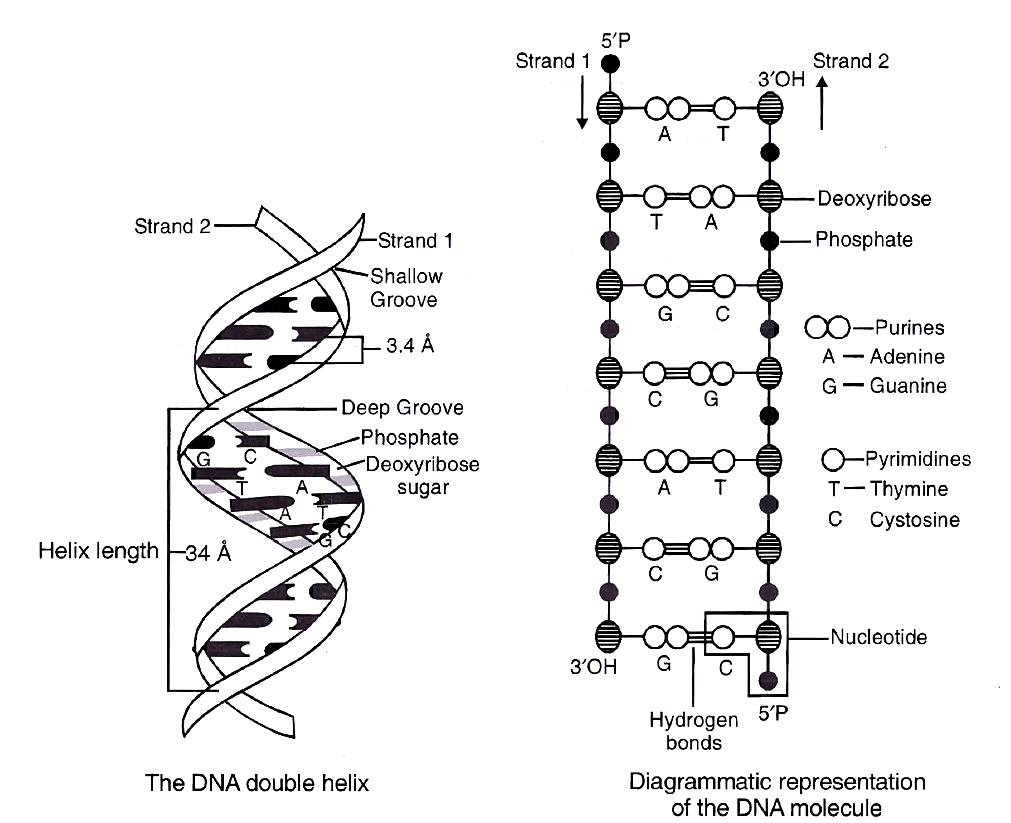
Concept Builder
Types of DNA and their comparison
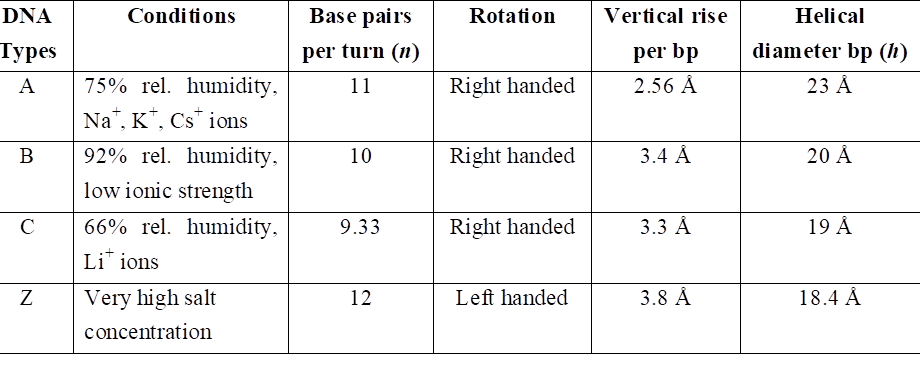
PACKAGING OF DNA HELIX
The average distance between two adjacent base pairs is 0.34 nm (0.34 × 10–9 m or 3.4 Å).
Length of DNA for a human diploid cell is 6.6 × 109 bp × 0.34 × 10–9 m/bp = 2.2 m.
This length is far greater than the dimension of a typical nucleus (approximately 10–6 m).
The number of base pairs in Escherichia coli is 4.6 × 106. The total length is 1.36 mm.
The long sized DNA is accommodated in small area (about 1 µm in E. coli) only through packing or compaction.
DNA is acidic due to presence of large number of phosphate group.
Compaction occurs by folding and attachment of DNA with basic proteins, polyamine in prokaryotes and histone in eukaryotes.
DNA packaging in Prokaryotes :
DNA is found in cytoplasm in supercoiled state.
The coils are maintained by non histone basic protein like polyamines.
RNA may also be involved. This compact structure of DNA is called nucleoid or genophore.
DNA packaging in Eukaryotes :
It is carried out with the help of lysine and ariginine rich basic proteins called histones.
The unit of compaction is called nucleosome.
There are five types of histone proteins H1, H2A, H2B, H3 and H4.
Four of them occur in pairs to produce histone octamer (2 copies of each -H2A, H2B, H3 and H4), called nu body or core of nucleosome.
Their positively charged ends are directed outside.
They attract negatively charged strands of DNA.
About 200 bp of DNA is wrapped over nu body to complete about 1¾ turns.
This forms a nucleosome of size 110 × 60 Å (11 × 6 nm).
DNA present between two adjacent nucleosome is called linker DNA.
It is attached to H1 histone protein.
Length of linker DNA varies from species to species.
Nucleosome chain gives a beads on string appearance under electron microscope.
The nucleosomes further coils to form solenoid.
It has diameter of 30 nm as found in chromatin.
The beads-on-string structure in chromatin is packaged to form chromatin fibres that are further coiled and condensed at metaphase stage of cell division to form chromosomes.
The packaging at higher level requires additional set of proteins (acidic) that collectively are referred to as non-histone chromosomal (NHC) proteins.
Non-Histone Chromosomal proteins are of three types:
(i) Scaffold or Structural NHC protein.
(ii) Functional NHC protein, e.g., DNA polymerase, RNA polymerase.
(iii) Regulatory NHC protein, e.g., HMG (High mobility group proteins that controls gene expression).
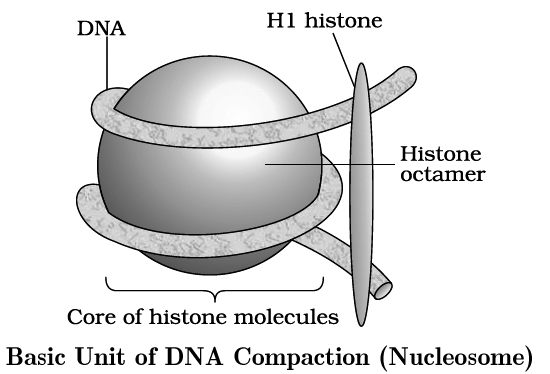
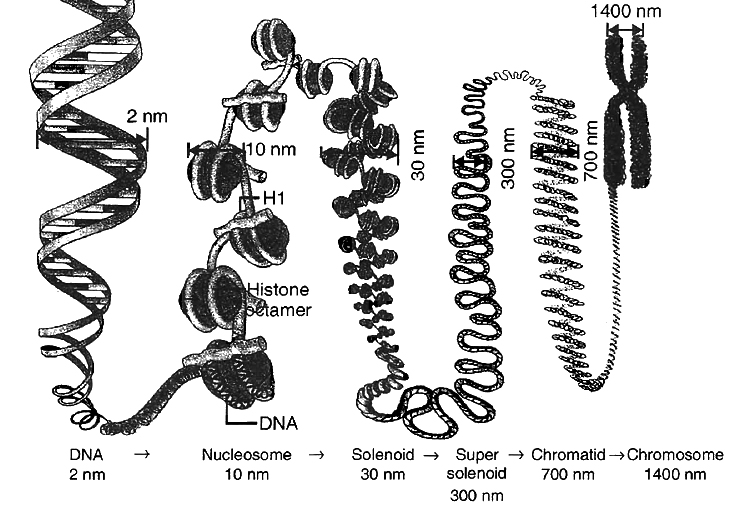
In a typical nucleus, some region of chromatin are loosely packed (and stains light) and are referred to as euchromatin.
The chromatin that is more densely packed and stains dark is called as heterochromatin, specifically euchromatin is said to be transcriptionally active and heterochromatin is transcriptionally inactive.
Chemical Composition of Chromosome
A chromosome consists of following chemical compositions:
(a) DNA 40%
(b) RNA 1.2%
(c) Histone protein 50%
(d) Acidic proteins 8.5%
(e) Lipid traces
(f) Ca+2, Mg+2, Fe+2 traces
the search of genetic material
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
THE SEARCH FOR GENETIC MATERIAL
The experiments given below prove that DNA is the genetic material.
(I) Evidence from bacterial transformation.
The transformation experiments, conducted by Frederick Griffith in 1928, are of great importance in establishing the nature of genetic material.
He used two strains of bacterium Diplococcus or Streptococcus pneumoniae or Pneumococcus i.e., S-III and R-II.
(1) Smooth (S) or capsulated type which have a mucous coat and produce shiny colonies. These bacteria are virulent and cause pneumonia.
(2) Rough (R) or non-capsulated type in which mucous coat is absent and produce rough colonies. These bacteria are nonvirulent and do not cause pneumonia.
The experiment can be described in following four steps :
(a) Smooth type bacteria were injected into mice. The mice died as a result of pneumonia caused by bacteria.
S strain Injected into mice Mice died
(b) Rough type bacteria were injected into mice. The mice lived and pneumonia did not occur.
R strain Injected into mice Mice lived
(c) Smooth type bacteria which normally cause disease were heat killed and then injected into the mice. The mice lived and pneumonia was not caused.
S strain (heat killed) Injected into mice Mice lived
(d) Rough type bacteria (living) and smooth type heat-killed bacteria (both known not to cause disease) were injected together into mice. The mice died due to pneumonia and virulent smooth type living bacteria could also be recovered from their dead bodies.
S strain (heat killed) + R strain (living) Injected into mice Mice died
He concluded from fourth step of the experiment that some rough bacteria (nonvirulent) were transformed into smooth type of bacteria (virulent).
This occurred perhaps due to absorption of some transforming substance by rough type bacteria from heat killed smooth type bacteria.
This transforming substance from smooth type bacteria caused the synthesis of capsule which resulted in production of pneumonia and death of mice.
Therefore, transforming principle appears to control genetic characters (for example, capsule as in this case). However, the biochemical nature of genetic material was not defined from his experiments.
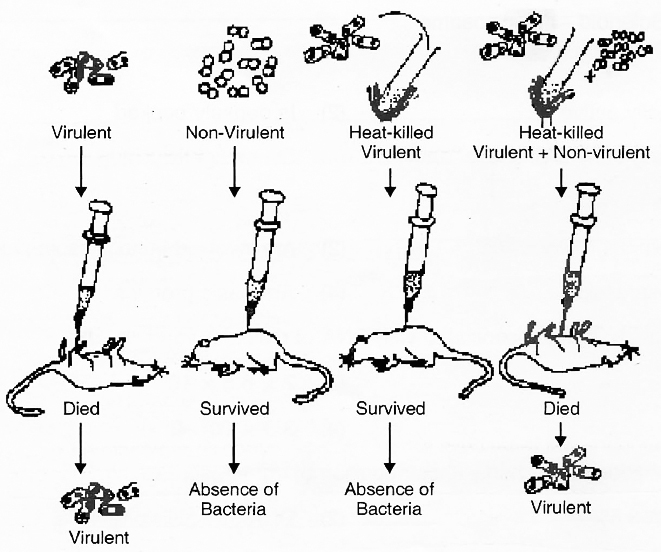
Biochemical Characterisation of Transforming Principle
Later, Avery, Macleod and McCarty (1944) repeated the experiment in-vitro to identify the biochemical nature of transforming substance. They proved that this substance is DNA.
Pneumococcus bacteria cause disease when capsule is present. Capsule production is under genetic control.
In the experiments, rough type bacteria (non-capsulated and non-virulent) were grown in a culture medium to which DNA extract from smooth type bacteria (capsulated and virulent) was added.
Later, the culture showed the presence of smooth type bacteria also in addition to rough type.
This is possible only if DNA of smooth type was absorbed by rough type bacteria which developed capsule and became virulent.
This process of transfer of characters of one bacterium to another by taking up DNA from solution is called transformation.
When DNA extract was treated with DNase (an enzyme which destroys DNA), transformation did not occur.
The transformation occurs when proteases and RNases were used. This clearly shows that DNA is the genetic material.
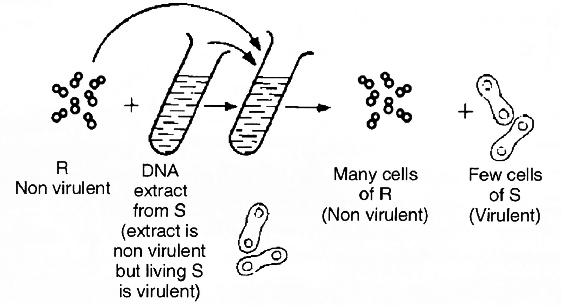
(II) Evidence from experiments with bacteriophage.
T2 bacteriophage is a virus that infects bacterium Escherichia coli and multiplies inside it.
T2 phage is made up of DNA and protein coat.
Thus, it is the most suitable material to determine whether DNA or protein contains information for the production of new virus (phage) particles.
Hershey and Chase (1952) demonstrated that only DNA of the phage enters the bacterial cell and, therefore, contains necessary genetic information for the assembly of new phage particle.
The functions of DNA and proteins could be found out by labelling them with radioactive tracers.
DNA contains phosphorus but not sulphur.
Therefore, phage DNA was labelled with P32 by growing bacteria infected with phages in culture medium containing 32P.
Similarly, protein of phage contains sulphur but no phosphorus.
Thus, the phage protein coat was labelled with S35 by growing bacteria infected with phages in another culture medium containing 35S.
After the formation of labelled phages. Three steps were followed, i.e., infection, blending, centrifugation.
1. Infection: Both types of labelled phages were allowed to infect normally cultured bacteria in separate experiments.
2. Blending: These bacterial cells were agitated in a blender to break the contact between virus and bacteria.
3. Centrifugation: The virus particles were separated from the bacteria by spinning them in a centrifuge.
After the centrifugation the bacterial cells showed the presence of radioactive DNA labelled with P32 while radioactive protein labelled with S35 appeared on the outside of bacteria cells (i.e., in the medium).
Labelled DNA was also found in the next generation of phage.
This clearly showed that only DNA enters the bacterial host and not the protein.
DNA, therefore, is the infective part of virus and also carries all the genetic information.
This provided the unequivocal proof that DNA is the genetic material.
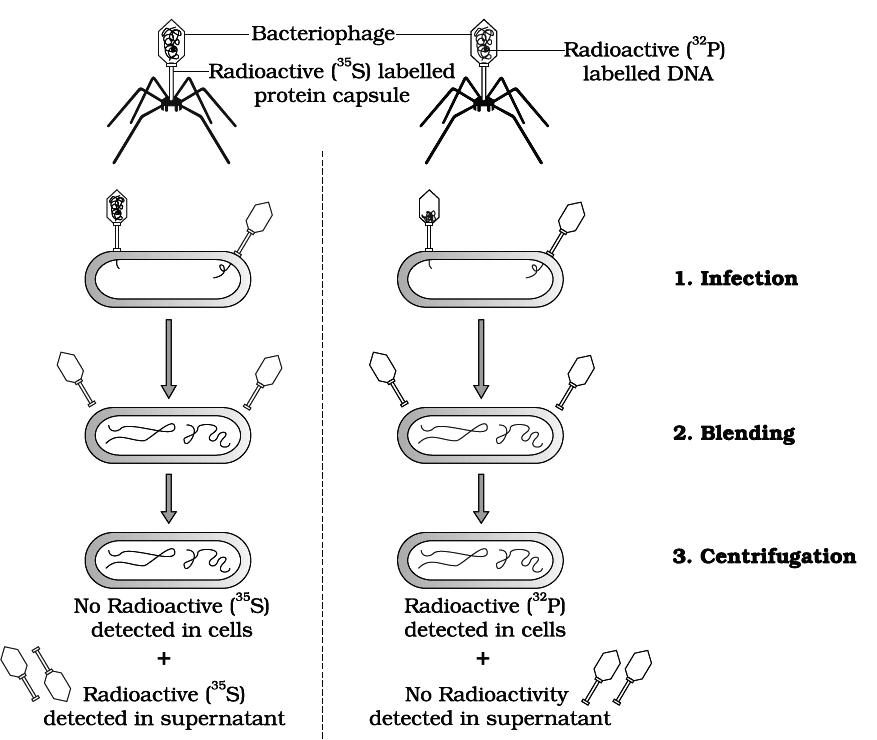
that DNA is genetic material or DNA is infective part of virus
Properties of Genetic Material :
Following are the properties and functions which should be fulfilled by a substance if it is to qualify as genetic material.
(1) It should chemically and structurally be stable.
(2) The genetic material should be able to transmit faithfully to the next generation, as Mendelian characters.
(3) The genetic material should also be capable of undergoing mutations.
(4) The genetic material should be able to generate its own kind (replication).
This can be concluded after examining the above written qualities, that DNA being more stable is preferred as genetic material, as
(a) Free 2OH of RNA makes it more labile and easily degradable. Therefore, DNA in comparison is more stable.
(b) Presence of thymine at the place of uracil also confers additional stability to DNA.
(c) RNA being unstable, mutates at a faster rate.
RNA world
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
RNA World
RNA was the first genetic material.
There are evidences to suggest that essential life processes, such as metabolism, translation, splicing, etc. evolved around RNA.
RNA used to act as a genetic material as well as a catalyst.
There are some important biochemical reactions in systems that are catalyzed by RNA catalysts and not by proteinaceous enzymes (e.g., splicing).
RNA being a catalyst was reactive and hence unstable .
Therefore, DNA has evolved from RNA with chemical modifications that make it more stable.
DNA being double stranded and having complementary strand further resists changes by evolving a process of repair.
RNA is an adapter, structural molecule and in some cases catalytic.
Thus, RNA is better material for transmission of information.
Dna replication
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
REPLICATION OF DNA
The Watson-Crick model of DNA immediately suggested that the two strands of DNA would separate.
Each separated or parent strand serves as a template (model or guide) for the formation of a new but complementary strand.
Thus, the new or daughter DNA molecules formed would be made of one old or parental strand and another newly formed complementary strand.
This method of formation of new daughter DNA molecules is called semi-conservative method of replication.
The following experiment suggests that DNA replication is semi-conservative.
Messelson and Stahl (1958) conducted experiments using heavy nitrogen (15N) to determine whether the concept of semi-conservative replication is correct.
They used Cesium chloride (CsCl) gradient centrifugation technique for this purpose.
A dense solution of CsCl, on centrifugation, forms density gradient-bands of a solution of lower density at the top that increases gradually towards bottom with highest density.
If DNAs of different densities are mixed with CsCl solution, these would separate from one another and would form a definite density band in the gradient along with CsCl solution.
Messelson and Stahl created DNA molecules of different densities by using normal nitrogen 14N and its heavy isotope 15N.
For this purpose, Escherichia coli was grown in 15NH4Cl containing culture medium for many generations, so that bacterial DNA become completely heavy.
This non-radioactive or heavy DNA (incorporating 15N) had more density than DNA with normal nitrogen (14N).
The bacteria were then transferred to culture medium containing only normal nitrogen (14NH4Cl). The change in density was observed by taking DNA samples periodically.
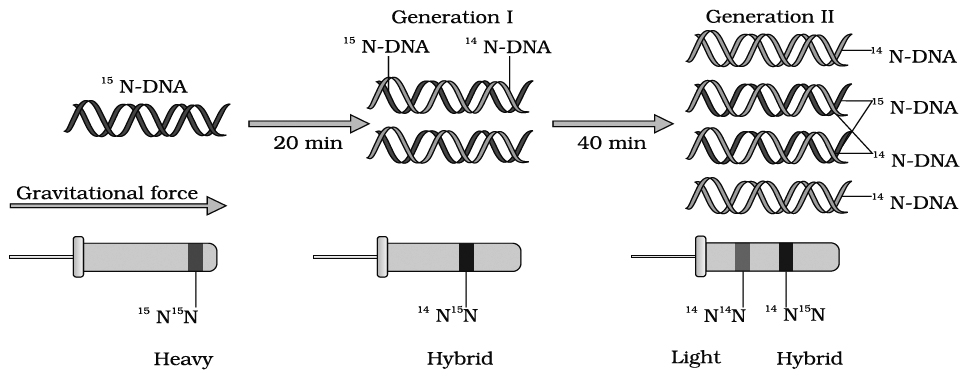
Messelson and Stahl's Experiment
If DNA replicates semi-conservatively, then each heavy (15N) DNA strands should separate and each separated strand should acquire a light (14N) partner after one round of replication.
This should be a hybrid DNA made of two strands, i.e., 14N–15N.
Meselson and Stahl observed that such DNA was actually half dense indicating the presence of hybrid DNA molecules.
After second round of replication there would be four DNA molecules.
Of these, two molecules would be hybrid (14N–15N) showing half density as earlier and the remaining two molecules would be made of light strands (14N–14N) Thus, after second generation, the same half dense band (14N–15N) was seen but the density of light bands (14N–14N) increased.
Messelson and Stahl's work as such provided confirmation of Watson-Crick model of DNA and its semi-conservative replication.
Taylor proved semiconservative mode of chromosome replication in eukaryotes using tritiated thymidine in the root of Vicia faba (faba beans).
Cairns proved semiconservative mode of replication in E. coli by using tritiated thymidine (H3–tdR) in autoradiography experiment.
He proposed -model for replication in circular DNA.
Mechanism of DNA Replication
DNA replication involves following four major steps:
(1) Initiation of DNA replication
(2) Unwinding of helix
(3) Formation of primer strand
(4) Elongation of new strand
1. Initiation of replication.
Replication of DNA always begins at a definite site called origin of replication.
Prokaryotes have single origin of replication.
It is called ori-c in E.coli.
On the other hand, eukaryotes have several thousand origins of replication.
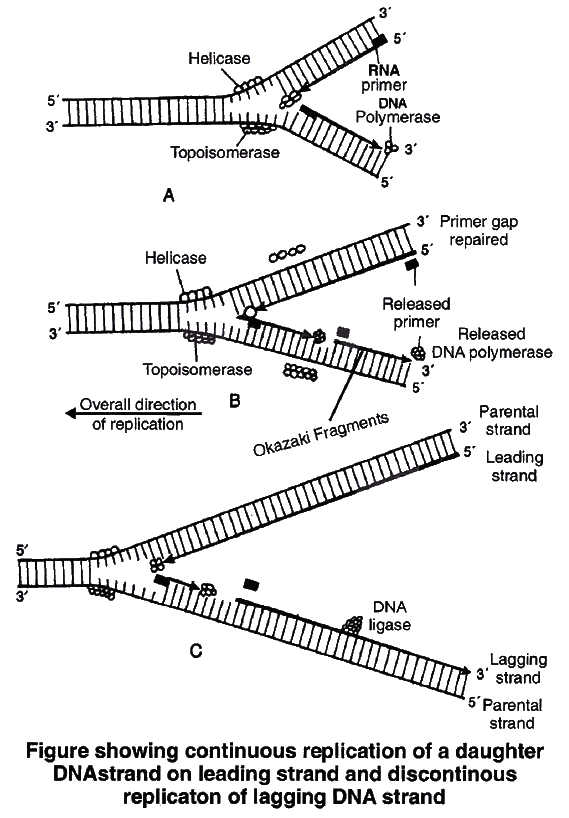
2. Unwinding of helix.
DNA replication requires that a double helical parental molecule is unwound so that its internal bases are available to the replication enzymes.
Unwinding is brought about by enzyme helicase which is ATP dependent.
Unwinding of DNA molecule into two strands results in the formation of Y-shaped structure, called replication fork.
These exposed single strands are stabilised by a protein known as Single-strand binding protein (SSB).
Due to unwinding, a supercoiling develops on the end of DNA opposite to replicating fork.
This tension is released by enzyme topoisomerase.
3. Formation of primer strand.
A new strand is to be synthesized opposite to the parental strands DNA polymerase III is the true replicase in E. coli, which is incapable of initiating DNA synthesis, i.e., it is unable to deposit the first nucleotide in a daughter (new) strand without the Primer.
Another enzyme, known as primase, strand synthesizes a short primer strand of RNA.
The primer strand then serves as a stepping stone (to start errorless replication).
Once the initiation of DNA synthesis is completed, this primer RNA strand is then removed enzymatically.
4. Elongation of new strand.
Once the primer strand is formed, DNA replication occurs in 53 direction, i.e., during synthesis of a new strand, deoxyribonucleoside triphosphates (dATP, dGTP, dTTP, dCTP) are added only to the free 3OH end.
Thus, the nucleotide at 3 end is always the most recently added nucleotide to the chain.
As the DNA replication proceeds on the two parental strands, synthesis of daughter or new strand occurs continuously along the parent 3'5' strand.
It is now known as leading daughter strand.
Synthesis of another daughter strand along the other parental strand, however, takes place in the form of short pieces.
This is called lagging daughter strand. These short pieces of DNA are known as Okazaki fragments, these segments are about 1,000-2,000 nucleotides long in prokaryotes.
Hense, DNA replication is semidiscontinuous. Discontinuous pieces of the lagging strand are joined together by the enzyme DNA ligase (after removal of primer) to form continuous daughter strand.
Thus two DNA molecules are now formed from one molecule.
Each of these daughter DNA molecules is made of two strands, of which one is old (parental) and other one is new or complementary strand.
DNA polymerase is the most important enzyme of DNA replication.
DNA polymerases are of three types i.e. DNA polymerase I, II and III in prokaryotes.

DNA polymerase II & III have only 3' 5' exonuclease activity.
Kornberg (1956), succeeded in demonstrating the in vitro synthesis of DNA molecule using a single strand of DNA as a template.
He extracted and purified an enzyme from E. coli which was capable of linking free DNA nucleotides, in presence of ATP as an energy source, to form complementary strand.
He called it DNA polymerase. DNA polymerase-I is called Kornberg enzyme.
In eukaryotes DNA polymerases are of 5 type these are DNA polymerase , , , and .
In eukaryotes, the replication of DNA takes place at s-phase of the cell cycle. The replication of DNA and cell division cycle should be highly co-ordinated. A failure in cell division after DNA replication result into polyploidy (achromosomal anomaly).
STRUCTURE OF RNA (Ribose Nucleic Acid)
RNA or ribonucleic acid is present in all the living cells. It is laevo rotatory and is responsible for learning and memory. It is found in the cytoplasm as well as nucleus. Description of types and structure of RNA is given below.
Types of RNA
In bacteria, there are three major types of RNAs: mRNA (messenger RNA), tRNA (transfer RNA), and rRNA (ribosomal RNA).
All three RNAs are needed to synthesise a protein in a cell.
The mRNA provides the template, tRNA brings aminoacids and reads the genetic code, and rRNAs play structural and catalytic role during translation.
RNA is generally involve in protein synthesis but in majority of plant viruses, it serves as a genetic material. Therefore, there are two major types of RNA-(A) genetic RNA and (B) non-genetic RNA.
(A) Genetic RNA -
Fraenkel-Conrat showed that RNA present in TMV (Tobacco Mosaic Virus) is a genetic material. Since then, it is established that RNA acts as a genetic material in most plant viruses.
(B) Non-genetic RNA.
This type of RNA is commonly present in cells where DNA is genetic material. Non-genetic RNA is synthesized on DNA template. It is of following three major types:
(a) Messenger RNA (mRNA).
It was reported by Jacob and Monad.
It carries genetic information present in DNA, so is called working copy.
mRNA constitutes about 3-5% of the total RNA present in the cell.
The molecular weight varies from 25,000 to 1,00,000.
It is about 300 nucleotide long at minimum.
Structure of prokaryotic m-RNA. It is polycistronic in nature i.e., several cistrons (functional part of DNA) form a single m-RNA. Thus, each m-RNA has a message to produce several polypeptides. Average life is 2 minutes.
At 5' end near initiation codon a sequence of fixed bases is found called Shine Dalgarno(SD) sequence (5'AGGAGGU3'). It helps in correct binding of ribosomal subunit (30 S) on it.
Structure of eukaryotic m-RNA. It is monocistronic in nature and has a message to produce on polypeptide only. They are metabolically stable and their life varies from few hours to days.

UTR (Untranslated region). They are sequences of RNA before start or initiation codon and after stop or termination codon. They are not translated. They are transcribed as part of same transcript as the coding region. Such UTRs provide stability to m-RNA and also increase translational efficiency.
(b) Ribosomal RNA (rRNA). It was reported by Kuntz. It is most stable type of RNA and is a constituent of ribosomes. It forms about 80% of the total cellular RNA.
In eukaryotes, 4 types of rRNA's found are 28 S, 18 S, 5.8 S and 5 S whereas, in prokaryotes three types of rRNA's found are 23 S, 16 S and 5 S. It is synthesised by genes present on DNA of several chromosomes found within a region known as nucleolar organiser.
(c) Transfer RNA (tRNA). The existence of tRNA was postulated by Crick. It is also known as soluble RNA (sRNA). These are the smallest molecules which carry amino acids to the site of protein synthesis. These constitute about 15% of the total cellular RNA.
It acts as an intermediate molecule between triplet code of mRNA and amino acid sequence of polypeptide chain. All tRNA's have almost same basic structure. There are over 60 types of tRNA.
Structure of t-RNA
Clover leaf model of tRNA is a 2-dimensional model suggested by Holley et.al. tRNA molecule appears like a clover leaf, being folded with three or more double helical regions (stem), having loops also. The three dimensional structure of the tRNA was proposed to be inverted L-shaped by Kim and Klug.
All tRNA molecules commonly have a guanine residue at its 5' terminal end. At its 3' end, unpaired – CCA sequence is present. Amino acid gets attached at this end only. The number of nucleotide varies from 77 (tRNAalanine) to 207 (tRNAtyrosine).
There are three loops in t-RNA
(i) Amino acyl synthetase binding loop, also called DHU loop.
(ii) Ribosomal binding loop with 7 unpaired bases. It is also called TC loop.
(iii) Anticodon loop with 7 unpaired bases. Out of the 7 bases in anticodon loop, 3 bases acts as anticodon for a particular triplet codon present on mRNA.
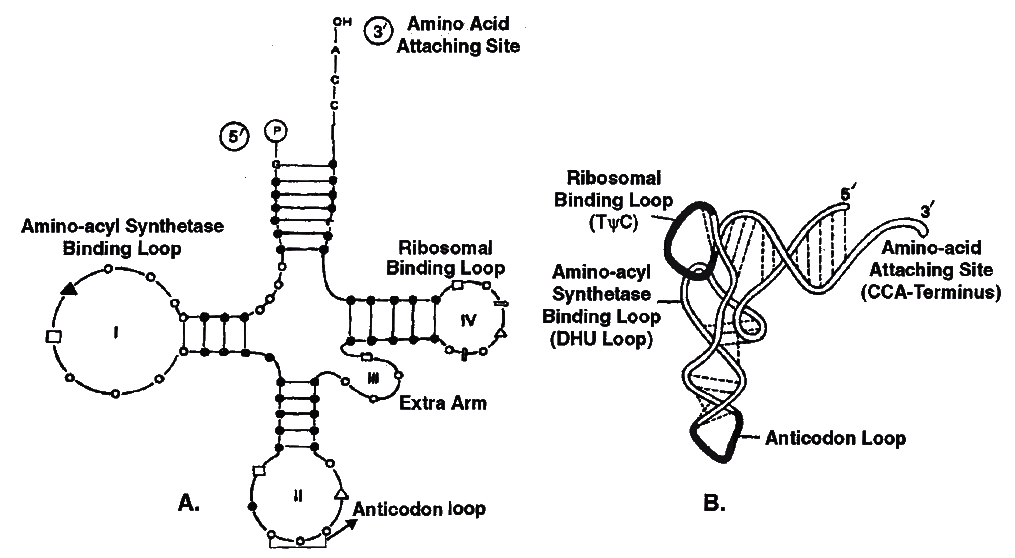
Structure of tRNA
A. Clover leaf model to show basic plan of tRNA secondary structure or 2D structure
B. Three Dimensional Structure showing L-shaped configuration
GENE EXPRESSION
DNA, being the genetic material, carries all the informations necessary to programme the functions of a cell by controlling the synthesis of enzymes or proteins.
Beadle and Tatum put forward a theory-one gene one enzyme in support of the earlier hypothesis that enzymes are proteinaceous in nature and each is produced by a single gene.
They conducted experiments on the nutritional strains of pink mould, Neurospora crassa.
This fungus grows on simple nutrient medium and has the ability to synthesize all its cellular components.
Such an organism is called prototroph.
An organism that is unable to synthesize a particular cellular metabolite, such as an amino acid or coenzyme is called auxotroph.
Beadle and Tatum produced arginine (an amino acid) auxotrophs (mutants of Neurospora unable to synthesize arginine) by giving X-rays treatment to the cells. Arginine synthesis passes through the following path

They found that any step of this metabolic chain could be blocked by a mutation in a specific enzyme catalyzing the reaction, each enzyme representing a different gene product.
Thus, Beadle and Tatum reached a conclusion that each gene functions to produce a single enzyme.
Some proteins, e.g., haemoglobin and other quaternary proteins are made up of two or more than two polypeptide chains. After it, the 'one gene one enzyme' theory was modified into one gene one polypeptide hypothesis by Yanofsky. Later Jacobson and Baltimore proposed one mRNA -one polypeptide hypothesis.
Gene and protein : A gene expresses itself by protein synthesis.
When a particular gene is expressed (i.e., controls a function or a reaction), its information is copied into another nucleic acid, i.e., mRNA (messenger RNA) which in turn directs the synthesis of specific proteins.
These concepts form the central dogma of molecular biology. This has been shown by F. Crick in the following diagram.
An exception to this one way flow of information was reported in 1970.
H. Temin and D. Baltimore independently discovered reverse transcription in some viruses.
These viruses can code an enzyme reverse transcriptase which can code DNA on RNA template.
This discovery was important in understanding cancer and, hence, these two scientists were awarded Nobel Prize. The modified flow of information now can be shown as below:

 Reverse flow of transcriptional information or Reverse Central Dogma or Teminism
Reverse flow of transcriptional information or Reverse Central Dogma or Teminism
Commoner suggests circular flow of information.
transcription
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
MECHANISM OF PROTEIN SYNTHESIS
The process of protein synthesis consists of two major steps:
(A) Transcription or synthesis of mRNA on DNA
(B) Translation or synthesis of proteins along mRNA
(A) Transcription
The transfer of genetic information from DNA to mRNA is known as transcription. The segment of DNA that takes part in transcription is called transcription unit. It has three components.
(i) A Promoter
(ii) The Structural gene
(iii) A Terminator
Promoter seguences are present upstream (5' end) of the structural genes of a transcription unit.
The binding sites for RNA polymerase lie within the promoter sequence.
Certain short sequences within the promoter sites are conserved. In prokaryotes, 10 bp upstream from, the start point lies a conserved sequence described as –10 sequence TATAAT or "Pribnow box" and –35 sequence TTGACA as "Recognition sequence".
Schematic structure of a transcription unit
Structural gene is part of that DNA strand which has 3' 5' polarity, as transcription occur in 5' 3' direction. This strand of DNA that direct the synthesis of mRNA is called template strand. The complementary strand is called non-template or coding strand, it is identical in base sequence to RNA transcribed from the gene, only with U in place of T.
Terminator is present at 3' end of coding strand and defines the end of the process of transcription.
Mechanism of transcription
RNA polymerase binds to promoter region of the DNA and the process of transcription begins.
RNA polymerase moves along DNA helix and unwinds it.
One of the two strands of DNA serves as a template for RNA synthesis.
This results in the formation of complementary RNA strand.
It is formed at a rate of about 40 to 50 nucleotides per second.
RNA synthesis comes to a stop when RNA polymerase reaches the terminator sequence.
The transcription enzyme, i.e., RNA polymerase is only of one type in prokaryotes and can transcribe all types of RNAs.
RNA polymerase is a holoenzyme that is represented as (2').
Molecular weight of holoenzyme is 4,50,000.
The enzyme without subunit is referred to as core enzyme.
Though, the core enzyme is capable of transcribing DNA into RNA but transcription starts non specifically at any base on DNA.
It is subunit which confers specificity. Rho factor () is required for termination of transcription.
(a) Sigma Factor () : Binds to the promoter site of DNA and it initiates transcription.
(b) Core Complex : It continues the transcription.
(c) Rho Factor () : It terminates transcription, its molecular weight is 55,000.
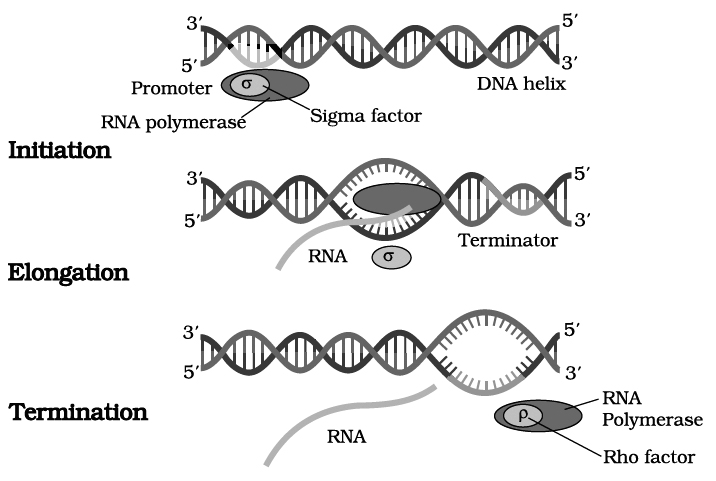
Transcription in Eukaryotes
In eukaryotes, the promotor site is recognised by presence of specific nucleotide sequence called TATA box or Hogness box (7 base pair long -TATATAT or TATAAAT) located 20 bp upstream to the start point.
Another sequence is CAAT box present between -70 and -80 bp.
In eukaryotes, the RNA polymerases are of three types, i.e., RNA polymerase I, RNA polymerase II and RNA polymerase III.
Functions of different RNA polymerases in eukaryotes are given below:
(i) RNA polymerase-I 5.8 S, 18 S, 28 S rRNA synthesis
(ii) RNA polymerase-II Hn RNA (heterogenous nuclear RNA), mRNA synthesis
(iii) RNA polymerase-III tRNA, Sc RNA (small cytoplasmic RNA), 5S rRNA and Sn RNA (small nuclear RNA) synthesis.
The gene in eukaryotes, however, is made of several pieces of base sequences coding for amino acids called exonic DNA, separated by stretches of non-coding sequences, commonly called intronic DNA.
Thus, the information on the eukaryotic gene for assembling a protein is not continuous but split.
This discovery is the result of works of Richard J. Roberts and Philip Sharp.
They shared 1993 Nobel Prize for Physiology and Medicine for the discovery of 'split genes' in higher organisms.
Most eukaryotic genes contain very long base sequences, all of which do not necessarily form mature mRNA.
The coding DNA sequences of the gene are called exons and the intervening non-coding DNA sequences are called introns.
All introns have GU at 5' end and AG at 3' end. Depending on the size of gene, the number and length of exons may vary from a few to more than fifty, alternating with stretches of DNA that contain no genetic information introns.
The nascent RNA synthesized by RNA polymerase-II is called hnRNA (heterogenous nuclear RNA).
It contains both unwanted base sequences (transcribed from introns) alternated with useful base sequences (transcribed from exons).
This primary transcript is converted into functional mRNA after post-transcriptional processing which involves 3 steps:
1. Modification of 5' end by Capping: Capping at 5' end occurs rapidly after the start of transcription.
The guanosine methylated at 7th position is added at 5' with the help of enzyme guanyl transferase.
Cap is essential for formation of mRNA-ribosome complex.
Translation is not possible if cap is lacking, because cap is identified by 18 S rRNA of ribosome unit.
2. Polyadenylation at 3' end (Tailing) : Poly (A) is added to 3' end of newly formed hn RNA with the help of enzyme Poly A polymerase. It adds about 200-300 adenyl ate residues.
3. Splicing of hnRNA (Tailoring) : In eukaryotes the coding sequences of RNA (exons) are interrupted by non coding sequences (introns).
Small nuclear RNA (snRNA) + Protein complex called small nuclear ribonucleo protein or SnRNPs (or snurps) play important role in this process.
Here the introns are removed and exons come in one plane.
This process is called splicing through which a mature mRNA is produced.
Normally, mRNA carries the codons of single complete protein molecule (monocistronic mRNA) in eukaryotes, but in prokaryotes, it carries codons from several adjacent DNA cistrons and becomes much longer in size (polycistronic mRNA).
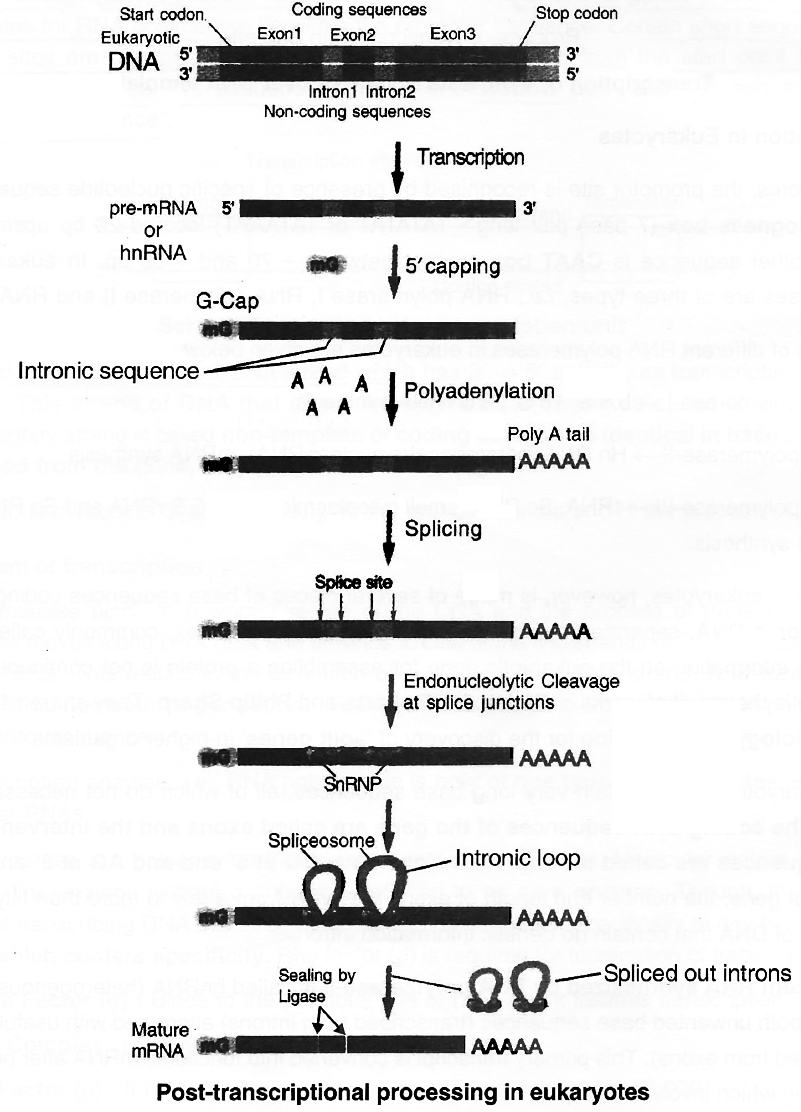
Genetic code
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
Genetic Code
Term was coined by Gamow.
DNA (or RNA) carries all the genetic information.
It is expressed in the form of proteins.
Proteins are made up of 20 different types of essential amino acids.
The information about the number and sequence of these amino acids forming protein is present in DNA and is passed on to mRNA during transcription.
Genetic code is a mRNA sequence containing coded information for one amino acid and consists of three nucleotides (triplet).
Thus, for twenty amino acids, 64 (4 × 4 × 4 or 43 = 64) minimum possible permutations are available.
This important discovery was the result of experiments by Marshall W. Nirenberg and J. Heinrich Matthaei and later by H.G. Khorana. Nirenberg and Khorana shared the 1968 Nobel Prize with R.W. Holley who gave details of tRNA structure. Nirenberg and Mathaei used a synthetic poly (U)RNA and deciphered the code by translating this as polyphenylalanine. Hargobind Khorana, using synthetic DNA, prepared polynucleotide with known repeating sequence, e.g., CUCUCUCUCUCU, it produced only two amino acids, leucine (CUC) and serine (UCU).
Properties of Genetic Code
1. Triplet code. Three adjacent nitrogen bases constitute a codon which specifies the placement of one amino acid in a polypeptide.
2. Start signal. Polypeptide synthesis is signalled by two initiation codons-AUG or rarely GUG. The first or initiating amino acid is methionine. The initiating codon on mRNA for methionine is AUG. Initiating methionine occurs in formylated state in prokaryotes. At other positions methionine is non-formylated. Both these methionines are carried by different tRNAs.
Rarely, GUG also serves as initiation codon. It normally codes for valine but if present at initiating position, it would code for methionine. So, GUG is an ambiguous codon .
3. Stop signal. Polypeptide chain termination is signalled by three termination codons-UAA (ochre), UAG (amber) and UGA (opal). They do not specify any amino acid and were hence called as nonsense codons. Whenever present in mRNA, these bring about termination of polypeptide chain and thus, act as stop signals. Codon UAA, UAG, UGA, AUG, and GUG are also called as punctuation codons.
4. Commaless. The genetic code is continuous and does not possess pauses (meaningless base) after each triplet. If a nucleotide is deleted or added, the whole genetic code will read differently. Thus, a polypeptide having 50 amino acids shall be specified by a linear sequence of 150 nucleotides. If a nucleotide is added or deleted in the middle of this sequence, the amino acids before this will be same but subsequent amino acids will be quite different.
5. Universal code. The genetic code is applicable universally, i.e., a codon specifies the same amino acid from a virus to human being or a tree.
6. Non-overlapping code. Every codon is independent and one codon does not overlap the next codon.
7. Degeneracy of code. Since there are 64 triplet codons and only 20 amino acids, the incorporation of some amino acids must be influenced by more than one codon. Only tryptophan and methionine are specified by single codons. All other amino acids are specified by 2-6 codons. The latter are called degenerate codons.
Wobble Hypothesis
A change in nitrogen base at the third position of a codon does not normally cause any change in the expression of the codon because the codon is mostly read by the first two nitrogen bases (wobble hypothesis of Crick).
The mutation that does not cause any change in the expression of the gene is called silent mutation.
The position of the third nitrogen base in a codon which does not influence the reading of the codon is termed as Wobble position.
It pairs with 1st position in anticodon. It means specificity of codon is determined by first two bases.
Wobbling helps one tRNA to read more than one codon and thus, provides economy in number of tRNA molecules at the time of translation.
Mutations and genetic code :
In a hypothetical mRNA, for example, the codons would normally be translated as follows :
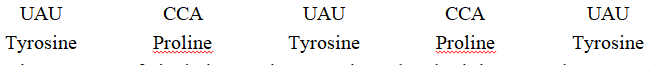
The insertion of single base G between the 3rd and 4th bases produces completely different protein from earlier one.

Similarly, the deletion of single base C at 4th place produces a new chain of amino acids and hence a different protein.

Such mutations are called frame shift mutations.Insertion or deletion of one or two nitrogenous bases changes the reading frame from the point of insertion or deletion.
However, insertion or deletion of three or its multiple bases insert or delete one or multiple codons hence one or multiple amino acids and reading frame remains unaltered from that point onwards.
This form genetic basis of proof that codon is a triplet and it is read in a contiguous manner.
Assignment of mRNA codons to Amino Acids
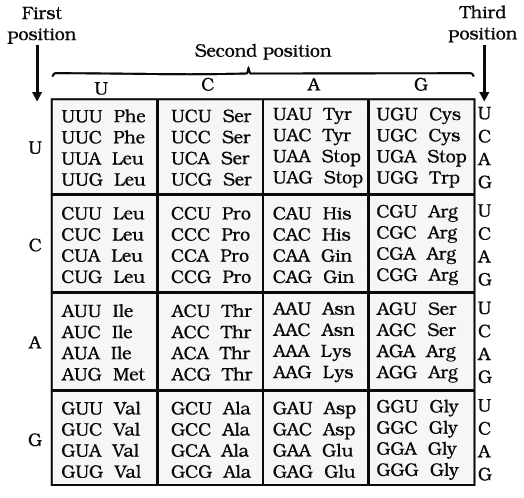
Translation
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
Translation
It is the mechanism by which the triplet base sequences of m-RNA molecules are converted into a specific sequence of amino acids in a polypeptide-chain. It occurs on ribosomes.
The major steps are:
(a) Activation of amino acids. In the presence of enzyme aminoacyl -tRNA synthetase (E), specific amino acids (AA) bind with ATP
![]()
 (b) Charging of t-RNA. The AA1-AMP-E1 complex formed in first step reacts with a specific tRNA. Thus, amino acid is transferred to tRNA. As a result, the enzyme and AMP are liberate
(b) Charging of t-RNA. The AA1-AMP-E1 complex formed in first step reacts with a specific tRNA. Thus, amino acid is transferred to tRNA. As a result, the enzyme and AMP are liberate
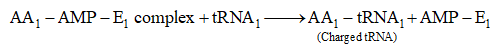
(c) Formation of polypeptide chain. It is completed in three steps.
(i) Chain Initiation. It requires 3 initiation factors in prokaryotes i.e., IF3, IF2, IF1.
9 initiation factors are required in eukaryotes i.e., eIF2, eIF3, eIF1, eIF4A, eIF4B, eIF4C, eIF4D, eIF5, eIF6.
(a) Binding of mRNA with smaller subunit of ribosomes (30S/40S). IF3 is involved in prokaryotes, while eIF2 is involved in eukaryotes. It involves interaction between Shine Delgarno sequence and 3' end of 16 S rRNA in prokaryotes.
(b) Binding of 30S / 40S-mRNA complex with t-RNA. Non-formylated methionine is attached with tRNA in eukaryotes and formylated methionine in prokaryotes. IF2 is involved in prokaryotes and eIF3 in eukaryotes.
![]()
(c) Attachment of larger subunit of ribosomes.
Initiation factors in eukaryotes -eIFl, eIF4.
Initiation factors in prokaryotes -IF1.
(ii) Chain elongation.
Ribosomes have two sites for binding amino acyl tRNA (i) Amino acyl or A site (acceptor site) (ii) Peptidyl site or P site (donor site).
A second charged tRNA molecule along with its appropriate amino acid approaches the ribosome at A site close to the P site.
Its anticodon binds to complementary codon of mRNA chain.
A peptide bond is formed between COOH group of first amino acid (methionine) and NH2 group of second amino acid.
The formation of peptide bond requires energy and is catalysed by enzyme peptidyl transferase. The initiating formyl methionine or methionine tRNA can bind only with P site.
All other newly coming aminoacyl tRNA bind to A site.
The elongation factor required for prokaryotes are EF-Tu and EF-Ts and in eukaryotes eEF1.
Translocation is movement of ribosome on mRNA. It requires EF -G in prokaryotes and eEF2 in eukaryotes.
(iii) Chain termination. The termination of polypeptide is signalled by one of the three terminal codons in the mRNA. The three terminal codons are UAG (Amber), UAA (Ochre) and UGA (Opal).
A GTP dependent factor known as release factor is associated with termination codon. It is eRF1 in eukaryotes and RF1 and RF2 in prokaryotes.
At the time of termination, the terminal codon immediately follows the last amino acid codon. After this, the polypeptide chain, tRNA, mRNA are released and the subunits of ribosomes get dissociated.
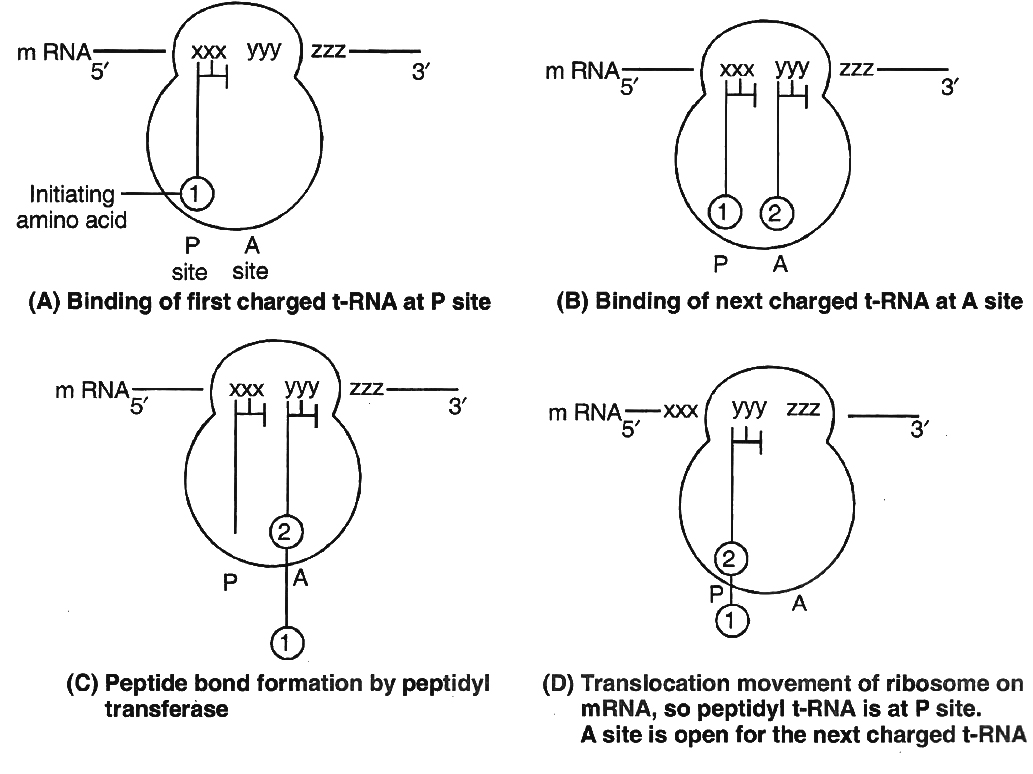
Concept Builder
There are some metabolic inhibitors which inhibit the synthesis of proteins in bacteria. These antibiotics are used in checking the growth of certain bacteria which are pathogenic in nature e.g.,
Tetracycline - Inhibits binding of amino-acyl tRNA to ribosomes
Streptomycin - Inhibits initiation of translation and causes misreading
Choloramphenicol - Inhibits peptidyl transferase activity
Erythromycin - Inhibits translocation of mRNA along ribosomes
Neomycin - Inhibits interaction between tRNA and mRNA.
Gene expression is the mechanism at the molecular level by which a gene is able to express itself in the phenotype of an organism.
In the translation unit, mRNA has some sequences that are not translated and are referred to as untranslated regions (UTR).
The UTRs are present at both 5' end (before start codon) and at 3' end (after stop codon).
They are required for efficient translation process.
Regulation of gene expression
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
REGULATION OF GENE EXPRESSION
1. Constitutive genes are those genes which are constantly expressing themselves in a cell because their products are required for the normal cellular activities, e.g., genes for glycolysis, ATPase.
2. Non-constitutive genes are not always expressing themselves in a cell. These are called Luxury genes. They are switched on or off according to the requirement of cellular activities, e.g., gene for nitrate reductase in plants, lactose system in Escherichia coli.
This provides maximum functional efficiency to the cell. Such regulated genes, therefore are required to be switched 'on' and 'off' when a particular function is to begin or stop.
This regulation can be achieved by anyone of the following two processes:
(1) Induction. This is a process of gene regulation where addition of a substrate stimulates or induces synthesis of enzymes needed for its own breakdown.
(2) Repression. In this process of gene regulation, addition of end product stops the synthesis of enzymes needed for its formation. This phenomenon is also known as feedback repression.
Operon
Francois Jacob and Jacques Monod (1961) proposed a model of gene regulation, known as Operon model.
Operon is a co-ordinated group of genes such as structural genes, operator gene, promoter gene, regulator gene which function or transcribe together and regulate a metabolic pathway as a unit.
Inducible operon (lac operon)
In Escherichia coli, breakdown of lactose requires three enzymes.
These enzymes are synthesized together in a co-ordinated manner and the unit is known as lac operon.
Since the addition of lactose itself stimulates the production of required enzymes, it is also known as inducible system.
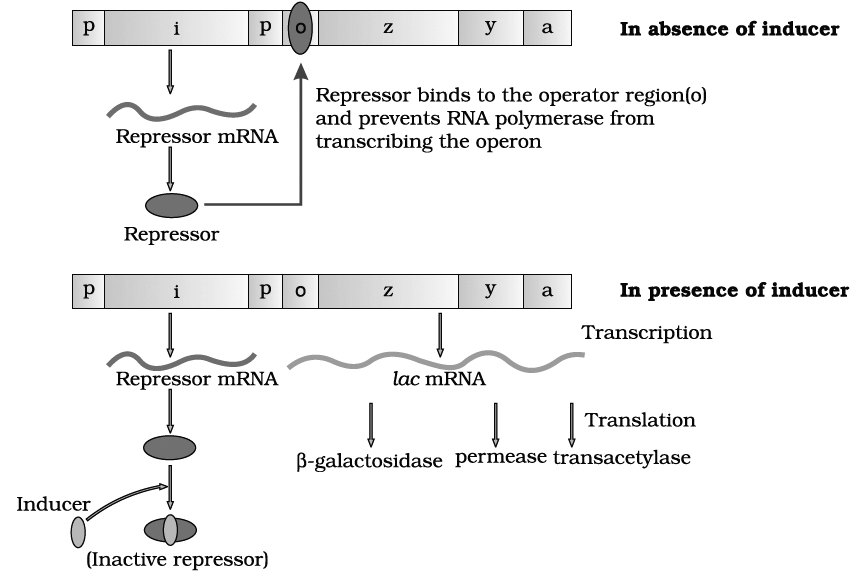
Lac operon
lac operon consists of following genes :
(1) Structural genes. These genes code for the proteins needed by the cell which include enzymes or other proteins having structural functions.
In lac-operon, there are following three structural genes:
(i) lac a -gene coding for enzyme transacetylase
(ii) lac y -gene coding for enzyme permease
(iii) lac z -gene coding for enzyme -galactosidase
(2) Operator gene (o). It interacts with a protein molecule (the regulator molecule), which promotes (induce) or prevents (repress) the transcription of structural genes.
(3) Promoter gene (p). This gene is the recognition point where RNA polymerase remains associated.
(4) Regulator gene (i). This is generally known as inhibitory gene (i). This gene codes for a protein called the repressor protein. It is an allosteric protein which can exist in two forms. In one form, it is paratactic to an operator and binds with it and in another form it is paratactic to inducer (like lactose).
The operon is switched 'off' and 'on'.
The transcription or function of lac genes or structural genes depends upon operator gene.
When repressor protein produced by inhibitory gene (i) or regulatory gene binds to operator (o) gene, RNA polymerase gets blocked. There would be no transcription and the operon model remains in 'switched off' position.
On the other hand, when inducer like lactose is added, the repressor protein (produced by gene i) gets bound to it.
The repressor, therefore, gets released from the operator.
RNA polymerase is now permitted to act.
Hence, the transcription of lac genes now takes place and the operon model is in 'switched on' position.
All the three genes are transcribed by RNA polymerase to form a single mRNA strand.
Each gene segment of mRNA is called cistron and, therefore, mRNA strand transcribed by more than one gene is known as polycistronic.
Tryptophan Operon -Repressible Operon System
Operon model can also be explained using feed-back repression.
In tryptophan (trp) operon, three enzymes are necessary for the synthesis of amino acid tryptophan.
These enzymes are synthesized by the activity of five different genes in a co-ordinated manner.
The addition of tryptophan, however, stops the production of these enzymes.
Thus, the system is known as repressible system.
In this system, there are five structural genes, trp A, trp B, trp C, trp D and trp E, coding for three enzyme needed for the synthesis of tryptophan, an amino acid.
Regulatory gene (R) produces repressor protein which is known as apo-repressor because it does not get bound to the operator directly.
Hence, the operator gene remains in 'switched on' position.
Tryptophan when added, binds to the apo-repressor and is called co-repressor.
This apo-repressor and co-repressor complex (activated repressor) now binds to operator gene and blocks the function of RNA polymerase.
Thus, transcription would not occur and tryptophan operon would be in 'switched off' position.
Feed-back repression is functional when there is no further need of the end product and, hence, there is no requirement of continuation of this anabolic pathway.
This operon stops the process.
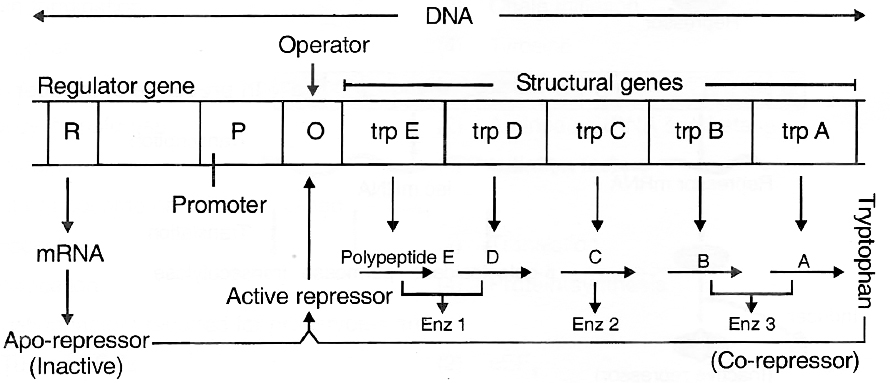
Tryptophan operon model of gene regulation in bacteria
In the prokaryotes, control of the rate of transcriptional initiation is the predominant site for control of gene expression.
Regulation of Gene Expression in Eukaryotes
In eukaryotes, the regulation of gene expression could be exerted at four levels:
(i) Transcriptional level (formation of primary transcript)
(ii) Processing level (regulation of splicing)
(iii) Transport of mRNA from nucleus to the cytoplasm
(iv) Translational level
In eukaryotes, functionally related genes do not represent an operon but are present on different sites in chromosomes.
Here, structural gene is called split gene which is a mosaic of exons and introns, i.e., base triplet -amino acid matching is not continuous.
Britten -Davidson gene battery model is most popular for eukaryotic genes.
It proposes the occurrence of 5-types of genes-producer, receptor, integrator, sensor and enhancer-silencer.
Exons are coding part of cistron that forms RNA.
Introns are non-translated part of DNA called IVS (intervening sequences) or spacer DNA.
An intron begins with GU and ends up with an AG.
However, the entire split gene is transcribed to form a continuous strip of hn-RNA.
This removal of non-coding intronic part and fusion of exonic coding parts of RNA is called RNA splicing. About 50-90% of primary transcribed RNA is discarded during processing.
Human genome project
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
Human Genome Project (HGP)
The Human genome project was the international, collaborative research program whose goal was the complete mapping and understanding of all the genes of human beings.
All genes together are known as "genome".
The Human Genome Project as "Mega project" was a 13 year project co-ordinated by the U.S. Department of Energy and the National Institute of Health.
An International Human Genome project was launched in the year 1990 and completed in 2003.
The International Human Genome sequencing consortium published the first draft of the human genome in the journal Nature in February 2001 .
Human genome is said to have approximately 3 × 109 bp, and the cost of sequencing required is US $ 3 per bp. The total estimated cost of the project would be approximately 9 billion US dollars. In human genome 20,000 to 25,000 genes are present, out of them smallest gene is TDF gene with 14 bp and largest gene is Duchenne Muscular Dystrophy gene with 2400 × 103 bp.
Organisms and the number of bp and genes in them
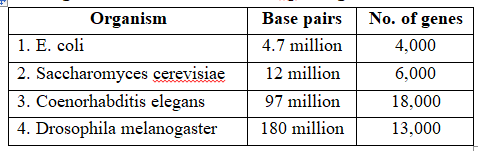
Following are the important goals of HGP :Goals of HGP
(i) Identication of all the approximately 20,000 -25,000 genes in human DNA.
(ii) To determine the sequence of the 3 billion chemical base pairs that make up human DNA.
(iii) To store this information in databases.
(iv) To improve tools for data analysis.
(v) Transfer related technologies to other sectors, such as industries.
(vi) Bioinformatics, i.e., close association of HGP with the rapid development of a new area in biology.
(vii) Sequencing of model organisms -Complete genome sequence of E.coli, Saccharomyces cerevisiae, Coenorhabditis elegans; Drosophila melanogaster, D. pseudoobscura, Oryza sativa and Arabidopsis was achieved in April, 2003.
(viii) Adress the ethical, legal and social issues (ELSI) that may arise from the project.
Concept Builder
Key Definitions :
1. cDNA: It stands for complementary DNA, a synthetic type of DNA generated from mRNA. By using mRNA as template, scientists use enzymatic reactions to convert its information back into cDNA and then clone it as cDNA library. These libraries are important to scientists because they consist of clones of all protein -encoding DNA or all the genes in the human genome.
2. Mb : Mb stands for megabase, a unit of length equal to 1 million base pairs and roughly equal to 1cM. (1cM = 1 million bp)
3. Microarray: Microarrays are devices used in many types of large scale genetic analysis. They can be used to study how large number of genes are expressed as messenger RNA in a particular tissue and how a cell's regulatory network control vast batteries of genes simultaneously.
Methodologies
The HGP techniques include:
(i) Sequence tagged site: It is a short DNA segment that occurs only once in a genome and whose exact location and order of bases is known. STSs serve as landmarks on the physical map of a genome. It is also called as Expressed Sequence Tags (ESTs). The genes that are expressed as RNA are referred to as ESTs.
(ii) Sequencing the whole set of genome that contained all the coding and non-coding sequences, and later assigning different regions in the sequence with functions, known as Sequence Annotation.
(iii) The Employment of Restriction Fragment Length Polymorphism (RFLP)
(iv) Yeast Artificial Chromosomes (YACs)
(v) Bacterial Artificial Chromosomes (BACs)
(vi) Polymerase chain reaction (PCR)
(vii) Electrophoresis
For sequencing, the total DNA from a cell is isolated and converted into random fragments of relatively smaller sizes (recall DNA is a very long polymer, and there are technical limitations in sequencin very long pieces DNA) and cloned in suitable host using specialised vectors.
The cloning resulted into amplification of each piece of DNA fragment so that it subsequently could be sequenced with ease.
The commonly used hosts were bacteria and yeast, and the vectors were called as BAC (bacterial artificial chromosomes), and YAC (yeast artificial chromosomes).
The fragments were sequenced using automated DNA sequencers that worked on the principle of a method developed by Frederick Sanger. These sequences were then arranged based on some overlapping regions present in them. This required generation of overlapping fragments for sequencing. Alignment of these sequences was humanly not possible. Therefore, specialised computer based programs were developed. These sequences were subsequently annotated and were assigned to each chromosome. The sequence of chromosome 1 was completed only in May 2006 (this was the last of the 24 human chromosomes -22 autosomes and X and Y -to be sequenced).
Salient Features of Human Genome
Some of the salient observations drawn from human genome project are as follows :
(i) The human genome contains 3164.7 million nucleotide bases.
(ii) The average gene consists of 3000 bases, but sizes vary greatly, with the largest known human gene being dystrophin at 2.4 million bases.
(iii) The total number of genes is estimated at 30,000-much lower than previous estimates of 80,000 to 1,40,000 genes. Almost all (99.9 percent) nucleotide bases are exactly the same in all people.
(iv) The functions are unknown for over 50 per cent of discovered genes.
(v) Less than 2 percent of the genome codes for proteins.
(vi) Repeated sequences make up very large portion of the human genome.
(vii) Repetitive sequences are stretches of DNA sequences that are repeated many times, sometimes hundred to thousand times. They are thought to have no direct coding functions, but they shed light on chromosome structure, dynamics and evolution.
(viii) Chromosome 1 has most genes (2968), and the Y has the fewest (231).
(ix) Scientists have identified about 1.4 million locations where single-base DNA differences (SNPs -single nucleotide polymorphism, pronounced as 'snips') occur in humans. This information promises to revolutionise the processes of finding chromosomal locations for disease-associated sequences and tracing numan history.
Classes of DNA sequences found in Human Genome
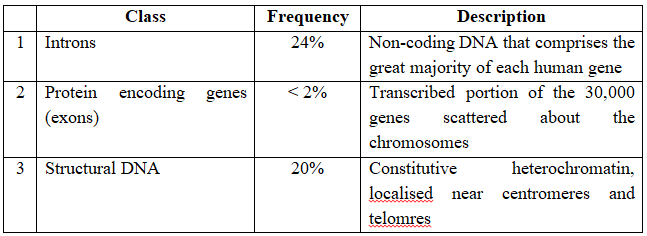
Francis Collins, director of the public funded genome project predicts the following progress in the next thirty years:Future Thrust of Human Genome Project
By 2010
1. Scientists will have developed accurate predictive tests for atleast a dozen common diseases so that preventive measures may be taken in advance.
2. Infertility specialists will be using the sophisticated techniques of pre-implantation genetic diagnosis to screen embryos for genetic disorders and designer babies.
3. Medicines will be making use of gene therapy.
By 2020
1. Doctors will have "designer drugs" to treat almost every disease.
2. Doctors will test patient's genetic make-up before prescribing drugs.
3. Doctors could change the genetic make-up of a living person by germ-line therapy.
4. Treatment of diseases like cancer, schizophrenia, depression etc. will have transformed.
By 2030
1. Genetic-based health care will have completely developed.
2. Most medical researches will be carried out using computer models rather than living tissues of animals.
3. Average life-span will rise up to 90 years.
The Indian Scenario
The Indian gene centre accounts for 160 species out of 2400 species in all of the 12 megagene centres.
Though the plant genetic resources activities got intensified in India in the first half of the century, but received the required impetus after creation of NBPGR in 1976 which has headquarters at IARI (Indian Agricultural Research Institute), New Delhi and several regional sub centres at different agroclimatic zones like Jodhpur (Arid areas), Trichur (Humid Tropical Zone), Shillong (Eastern sub tropical/subtemperate zone) etc.
As on 31 st March 1996, NBPGR holds 15,4,533 accessions of various agri-horticultural crops.
National Facility for Plant Tissue Culture Repository (NFPTCR) was established in 1986 by department of biotechnology at NBPGR for conservation of germplasm of vegetatively propagated plants.
Concept Builder
1. In translation, ribosomes are often found in clusters linked together by a strand of mRNA. This complex (polyribosome or polysome) enables several molecules of the same polypeptide to be produced simultaneously.
2. In vitro synthesis of gene was made by Hargobind Khorana (1968). He was able to link 77 nucleotide base pairs to code for yeast tRNAalanine. Later on in 1976, he synthesized a functional gene by linking 207 base pairs to code for E.coli. tRNA tyrosine.
3. Jumping genes (Transposon or Transposable element) discovered by Babara McClintock in maize can change their position within or between two chromosomes.
4. In some viruses a part of genetic material function for two or more genes (cistrons). Such genes are called overlapping genes. They were discovered by Borrel in × 174.
5. Transgenosis is the transmission of genetic information to plant cells by bacteriophage or phenomenon of transfer, gene maintenance, transcription and function.
6. Siblings (also sibship or sibs) : Individuals having the same parents; brother-sister relationship.
7. Genetic marker. A gene mutation that has phenotypic effects useful for tracing the chromosome on which it is located.
8. Phenocopy: An individual who has a phenotype similar to that produced by a certain mutant genotype, even though the individual may not have that genotype.
9. Charles Weismann isolated the gene (cDNA) for interferon from human lymphocytes and cloned it in E. coli and yeast.
10. Some Other Important Contributions of Scientists
(i) Zacharis - Called nuclein as DNA and proved that chromatin is rich in DNA
(ii) Fuelgen - DNA is located in nucleus where chromosomes are found, nucleus is fuelgen positive
(iii) Behrem - Nucleic acid is of two types DNA and RNA.
(iv) Fischer - Identified purines and pyrimidines in nucleic acid.
(v) Levene - Studied chemistry of DNA and found that DNA has deoxyribose sugar and four types of nucleotides.
(vi) Sanger - Determined base sequence in nucleic acid.
(vii) Hedges and Jacob -Term "transposon"
DNA fingerprinting
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
DNA FINGERPRINTING
What is DNA Fingerprinting?
The chemical structure of everyone's DNA is the same.
The only difference between people (or any animal) is the order of the base pairs.
There are so many millions of base pairs in each person's DNA that every person has a different sequence.
Using these sequences, every person could be identified solely by the sequence of their base pairs.
However, there are so many millions of base pairs due to which the task would be very time consuming.
Instead, scientists are able to use a shorter method, because of repeating base patterns in DNA (Satellite DNA).
These patterns do not, however, give an individual "fingerprint," but they are able to determine whether two DNA samples are from the same person, related people, or non-related individuals.
VNTR's, RFLP, SSR, RAPD
Every strand of DNA has stretches that contain genetic information which are responsible for an organism's development (exons) and stretches that, apparently, supply no relevant genetic information at all (introns).
Although the introns may seem useless, it has been found that they contain repeated sequences of base pairs.
These sequences are called Variable Number Tandem Repeats (VNTRs).
VNTR's also called minisatellites were discovered by Alec Jeffreys et. al. of U.K.
These consist of hypervariable repeat regions of DNA having a basic repeat sequence of 11-60 bp and flanked on both sides by restriction sites.
The number of repeats show a very high degree of polymophism and the size of VNTR varies from 0.1 to 20 Kb. The length of minisatellites and position of restriction sites is different for each person.
Therefore, when the genomes of two people are cut using the same restriction enzyme, the length and number of fragments obtained is different for both.
This is called RFLP or Restriction Fragment Length Polymorphism.
These fragments, when separated by gel electrophoresis, and obtained on a Southern Blot, constitutes what is called DNA fingerprint.
Father of DNA fingerprinting is Alec Jeffreys while the Indian experts Lalji Singh and V.K. Kashyap are known as Father of Indian technique.
Concept Builder
Now-a-days RAPD's or Randomly Amplified Polymorphic DNA are used for DNA fingerprinting (pronounced 'rapid').
It is a simpler technique than RFLP.
The DNA sequences flanking microsatellites (or SSRs i.e., simple sequence repeats or short tandem repeats) on both sides are conserved, so one can select primers complementary for these sequences and put these along with the genome in a thermal cycler.
The PCR will amplify the intervening microsatellites which can also be radiolabelled by using 32p.
The microsatellites have simple sequences of 1-6 bp. repeated hundreds of times.
Methodology of DNA fingerprinting
The Southern Blot is one way to analyze the genetic patterns which appear in a person's DNA. It was devised by E.M.Southern (1975) for separating DNA fragments. Performing a Southern Blot involves:
1. Isolating the desired DNA. It can be done either chemically, by using a detergent to wash the extra material from the DNA, or mechanically, by applying a large amount of pressure in order to "squeeze out" the DNA.
2. Cutting the DNA into several pieces of different size. This is done using one or more restriction enzymes.
3. Sorting the DNA pieces by size. The process by which the size separation is done is called gel electrophoresis. The DNA is poured into a gel, such as agarose, and an electrical charge is applied to the gel, with the positive charge at the bottom and the negative charge at the top. Because DNA has a slightly negative charge, the pieces of DNA will be attracted towards the bottom of the gel. The smaller pieces, however, will be able to move more quickly and thus, further towards the bottom than the larger pieces. The different-sized pieces of DNA will therefore, be separated by size, with the smaller pieces-towards the bottom and the larger pieces towards the top.
4. Denaturing the DNA fragments, so that all of the DNA is rendered single-stranded. This can be done either by heating or chemically treating the DNA in the gel using alkali.
5. Blotting the DNA. The gel with the size-fractionated. DNA is applied to a sheet of nitrocellulose paper or nylon membrane, and then baked to permanently attach the DNA to the sheet. The Southern Blot is now ready to be analyzed.
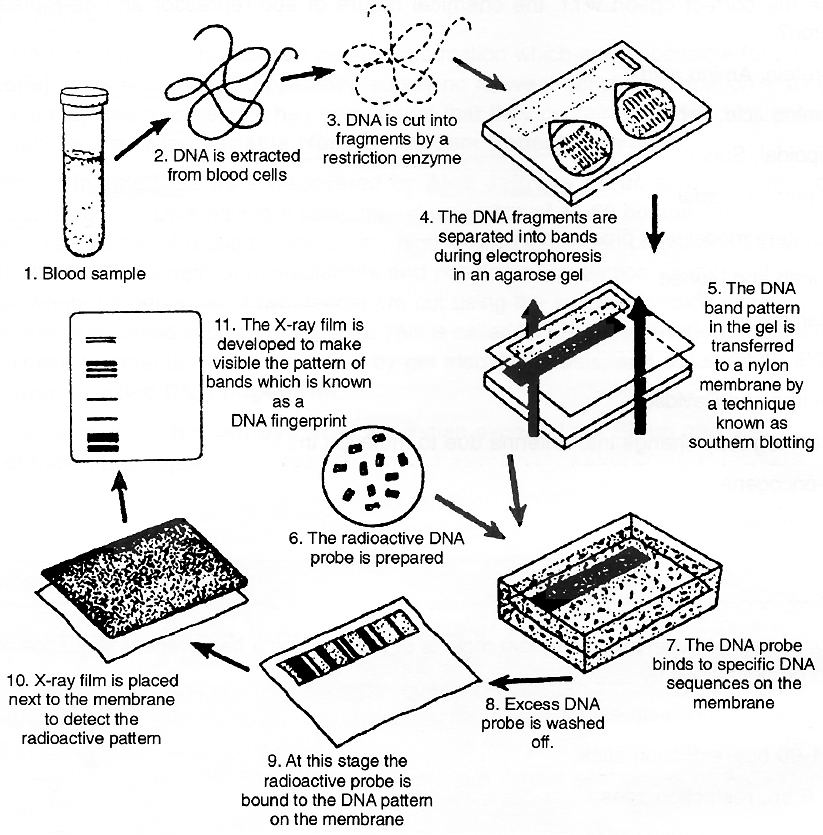
The DNA fingerprinting process
In order to analyze a Southern Blot, a radioactive genetic probe is used in a hybridization reaction with the DNA in question. If an X-ray is taken of the Southern Blot after a radioactive probe has been allowed to bound with the denatured DNA on the paper, only the areas where the radioactive probe binds will Show themselves on the film (autoradiography). This allows researchers to identify, in a particular person's DNA, the occurrence and frequency of the particular genetic pattern contained in the probe.
Practical Applications of DNA Fingerprinting
1. Solving Cases of Disputed Paternity and Maternity: Because a person inherits his or her VNTRs from his or her parents, VNTR patterns can be used to establish paternity and maternity.
2. Criminal Identification and Forensics: DNA isolated from blood, hair, skin cells, or other genetic evidence left at the scene of a crime can be compared, through VNTR patterns, with the DNA of a criminal suspect to determine guilt or innocence.
3. Personal Identification : The notion of using DNA fingerprints as a sort of genetic bar code to identify individuals has been discussed, but this is not likely to happen anytime in the near future. The technology required to isolate, keep on file, and then analyze millions of very specified VNTR patterns is both expensive and impractical.
GENOMICS
Application of DNA sequencing and genome mapping to the study, design and manufacture of biologically important molecules is known as genomics.
It is comparatively more recent branch in the field of biology.
The term genomics was introduced by Thomas Roderick.
In genomics, function of gene is identified by using the technique of reverse genetics.
Genomics study may be classified into three classes:
(i) Structural genomics : It involves mapping and sequencing of genes.
(ii) Functional genomics : It involves the identification of function of a particular gene.
(iii) Application genomics : It involves use of genomics information for crop improvement etc.
Complete genomics of Arabidopsis and rice have been worked out and they are having 130 and 430 million base pairs respectively.
How did life originate?
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
From Origin of Earth to Origin of Life
1. Evolution is a slow, continuous and irreversible process of change.
2. Origin of Earth: The Big Bang Theory proposes that the universe had an explosive beginning. The universe originated about 20 billion years ago by a big bang (thermonuclear explosion) of a dense entity. About 4.6 billion years ago, our solar system was probably created when the gaseous cloud called Solar Nebula started to collapse under the force of its own gravity, until it became a flattened spinning disc of atoms and particles. Its central region heated up and became a star.
3. Earth is about 4.6 billion years old, and the oldest rocks that have persisted in recognizable form are about 3.8 billion years old. For many years, scientists believed that such ancient rocks did not contain any fossils, but they knew that fossils were simply too small to be seen clearly without an electron microscope.
4. The oldest microfossils discovered so far are that of cyanobacteria that appeared 3.3 to 3.5 billion years ago.
5. Massive limestone deposits called Stromatolites became frequent in the fossil record about 2.8 billion years ago. Produced by cyanobacteria, stromatolites were abundant in virtually all freshwater and marine communities until about 1.6 billion years ago.
6. The fossil records indicate that unicellular protists -the first eukaryotes -appeared about 1.5 billion years ago.
7. Basic unit of evolution is population.
According to recent literature, the first non-cellularforms of life could have originated 3 billion years back. They would have been giant molecules (RNA, protein, polysaccharides etc.) These capsules reproduced their molecules perhaps. The first cellular form of life did not possibly originate till about 2000 million years ago.
Evolution of Life forms - A Theory
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
THEORIES ON THE ORIGIN OF LIFE
1. Theory of special creation states that the life was created by supernatural power in the form which has not undergone any change. It was given by Father Suarez. God created life in six days from materia prima and man was created by Him on the sixth day. According to this theory, earth is about 4000 years old.
2. Theory of catastrophism was given by Cuvier, according to which after a gap of certain period ( called age), the world undergoes a catastrophe (sudden calamity) which kills almost all the living organisms and then God creates a new generation or new life from inorganic matter.
3. Theory of biogenesis (i.e. life from life, omnis vivum ex. vivo) was proved by Redi, Spallanzani and Pasteur independently. They disproved (refuted) theory of spontaneous generation (abiogenesis). Francesco Redi (1668) proved that flies could not arise from putrefying meat without their eggs. Spallanzani (1767) demonstrated that putrefaction of meat is due to microbes in the air and it can be prevented by boiling and sealing the meat in air tight containers. Pasteur gave a definite proof of life arising from pre-existing life using microbes and sterilization methods. He performed "swan neck flask" experiment.
4. Cosmozoic theory (Theory of panspermia) given by Richter (1865); Helmholtz (1884), Arrhenius (1908) suggested that life reached the earth from some heavenly body through meteorites. Panspermia (primitive form of life, as suggested by Arrhenius, 1908) consisted of spores or seeds (sperms) and microbes that existed throughout universe and produced different forms of life on this earth.
Abiogenic or Chemical Origin of Life
Majority of the scientists are ofthe opinion that life originated from inanimate matter. Since the theory of abiogenic origin or chemical evolution of life is the only one that provides an explanation, which can be tested, most scientists have tentatively accepted it.
Oparin-Haldane Hypothesis
1. Alexander I. Oparin (1894-1980), a Russian biochemist, and J.B.S. Haldane (1892-1964), a British scientist, put forward the concept that the first living organism evolved from non-living material. They also suggested that the sequence of events that might have occured. In 1923, Oparin postulated that life originated on Earth at some point of time in the remote past, and under the conditions no longer observed. In his book, The Origin of Life (1938), Oparin submitted" abiogenesis first, butbiogenesis eversince". Oparin's theory is known as primary abiogenesis.
2. According to Oparin and Haldane (1929), spontaneous generation of early molecules might have taken place ifthe earth once had more reducing atmosphere compared to the present oxidising atmosphere. Oparin and Haldane agreed that the primeval Earth contained little, if at all, oxygen. Perhaps, in the primitive atmosphere oxygen in the free gaseous state was virtually absent. Therefore, no degradation of any organic compound arising in the primeval Earth could have taken place.
3. As there was no ozone layer in the atmosphere, any absorption of UV radiations, that is lethal to our present lives, was not possible in the primeval Earth.
4. The early gas cloud was rich in hydrogen, being present in the combined form in methane (CH4), ammonia (NH3) and water vapour (H2O).
5. Moreover, the atmospheric water vapour along with early gas cloud condensed into drops of water and fell as rain that rolled down the rock surfaces and accumulated to form liquid pools and oceans. In the process, erosion of rocks and washing of minerals (e.g., chlorides and phosphates) into the oceans were inevitable. Thus, Haldane's hot dilute soup was produced and the stage was set for combination of various chemical elements.
6. Atmospheric chemicals and those in water produced small precursor molecules, like amino acids, sugars, nitrogenous bases etc. These precursor molecules then combined resulting in the appearance of proteins, polysaccharides and nucleic acids.
7. The energy sources for such reactions of organic synthesis were the UV radiations (solar radiation), cosmic rays, electrical discharges (lightning), intense dry heat (volcanic eruption) and radioactive decay of various elements on the Earth's surface. Once formed, the organic molecules accumulated in water because their degradation was extremely slow in the absence of any life or enzyme catalysts. Such transformation is not possible in the present oxidising atmosphere because oxygen or micro organisms will decompose or destroy the living particle that may arise by mere chance.
Experimental Evidence for Abiogenic Molecular Evolution of Life
Harold C.Urey (1893 -1981), an astronomer, accorded the first adequate recognition of Oparin-Haldane's view on the origin of life in 1952.
Urey asked his student Stanely L. Miller, a biochemist, to replicate the primordial atmosphere as propounded by Oparin and Haldane.
Miller (1953) made the first successful simulated experiment to assess the validity of the claim for origin of organic molecules in the primeval Earth's conditions.
Concept Builder
1. Mllier sealed in a spark chamber a mixture of water (H2O), methane (CH4), ammonia (NH3) and hydrogen gas (H2). He made arrangement for insertion of two electrodes to provide electrical energy (simulation of lightning) to the spark chamber. CH4, NH3, H2 were in 2 : 1 : 2 ratio and water vapour at 800ºC. Electric sparks of 75,000 volt was provided to the mixture.
2. The spark chamber was connected to another flask with arrangement for boiling water (provision for evaporation). The other end of the spark chamber was connected to a trap by a tube that passed through a condenser (an arrangement for condensation and collection of aqueous solution, equivalent to rain and Haldane's soup). The trap, in turn, was connected with the flask for boiling water (arrangement for circulation).
3. The control apparatus contained every arrangement except that it was devoid of energy source.
4. After eighteen days, significant amount of the simple organic compounds (monomers), such as amino acids and peptide chains began to appear in the aqueous sample of the experimental set. Amino acids found were alanine, glycine and aspartic acid. Therefore, the obvious inference was that abiotic synthesis of organic monomers occurred in the simulated experimental condition. By analogy, such synthesis could have occurred in the primitive atmospheric condition. Later on many scientists repeated Miller's experiment using slightly different starting materials and UV radiation or other energy sources. All of them could successfully synthesise amino acid and related compounds. With hydrogen cyanide (HCN), even adenine and other nitrogen bases were produced.
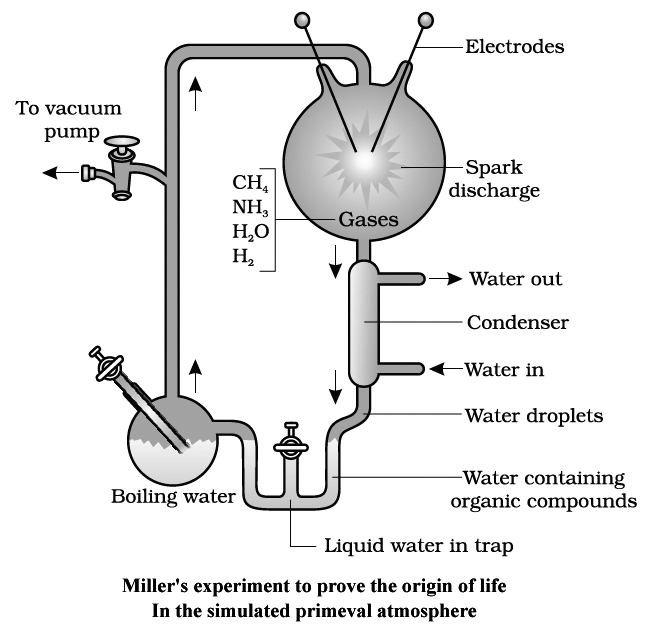
Abiotic synthesis of biomolecules is studied under following headings:
1. Chemogeny: Synthesis of organic molecules by chemical reactions.
2. Biogeny: Formation of self replicating biomolecules in broth (primordial hot soup or warm little pond).
3. Cognogeny: Evolution of various forms of life or diversification of existing groups.
Enclosing the Prebiotic Systems
The experiments of Miller and other scientists demonstrate that prebiotic molecules could have been formed under the conditions which most likely existed on early Earth.
Still, the formation of prebiotic soup of small molecules does not necessarily lead to the origin of life.
For origin of life, atleast three conditions needed to have been fulfilled:
1. There must have been a supply of self-replicators i.e., self-producing molecules.
2. Copying of these replicators must have been subject to error via mutation.
3. The system of replicators must have required a perpetual supply of free energy and partial isolation from the general environment.
The high temperature prevailing in early Earth would have easily fulfilled the second condition, that is, the requirement of mutation. The thermal motion would have continually altered the prebiotic molecules.
Concept Builder
The third condition, partial isolation, has been attained within aggregates of artificially produced prebiotic molecules.
These aggregates called protobionts can separate combinations of molecules from the surroundings; maintain an internal environment but are unable to reproduce.
Two important protobionts are coacervates and microspheres.
Oparin (1924) observed that if a mixture of a large protein and a polysaccharide is shaken, coacervates form.
Their interiors, which are primarily protein and polysaccharide, with some water, become separated from the surrounding aqueous solution.
The later has much lower concentration of proteins and polysaccharide.
Oparin's coacervates also exhibits a simple form of metabolism.
As these coacervates do not have lipid outer membranes and cannot reproduce, they fail to fulfil the requirement as a candidate of probable precursors of life.
Microspheres were formed when mixtures of artificially produced organic compounds were mixed with cool water.
If the mixture contains lipids, the surface of the microspheres consists of a lipid bilayer, reminiscent to the lipid bilayer of cell membranes.
Sydney Fox (1950) obtained protenoid microspheres.
There is considerable discussion among biologists as to how the first cells may have evolved.
The discovery made in the 1980's that RNA can act like enzyme to assemble new RNA molecules on an RNA template raised the interesting possibility that Coacervates may not have been the first step in the evolution of life.
Perhaps the first macromolecules were RNA molecules, and the initial steps on the evolutionary line were ones leading to more complex and stable RNA molecules.
Later, the stability might have been improved by surrounding the RNA within a coacervate.
Still other scientists reject the notion of 'RNA world', entirely, pointing out that some RNA components are too complex to have been present on the primitive earth.
Evidences of evolution
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
EVIDENCES OF EVOLUTION
1. EVIDENCES FROM ANATOMY
(i) Homologous organs :
These organs have the similar basic structure and developmental origin.
The organisms which possess such organs are said to have originated from common ancestor.
Consider, for example, the seal's flipper, the bat's wing, the horse's foot, the cat's paw and the human hand.
In plants, a thorn of Bougainvillaea differs from a tendril of Cucurbita in its function, both are located in a similar (axillary) position and have similar origin.
Thorns and tendrils are considered homologous.
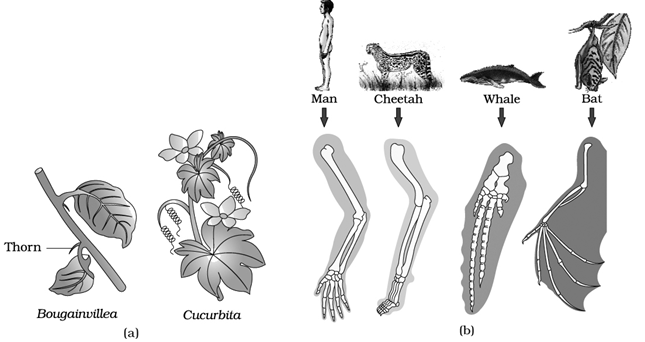
Homologous organs show divergent evolution which means that the similar structures developed along different directions due to adaptions to different needs or Adaptive Radiation which is the development of dissimilar functional structures in closely related group of organisms.
(Homology indicates common ancestry. Other examples are comparison of heart and brain of vertebrates)
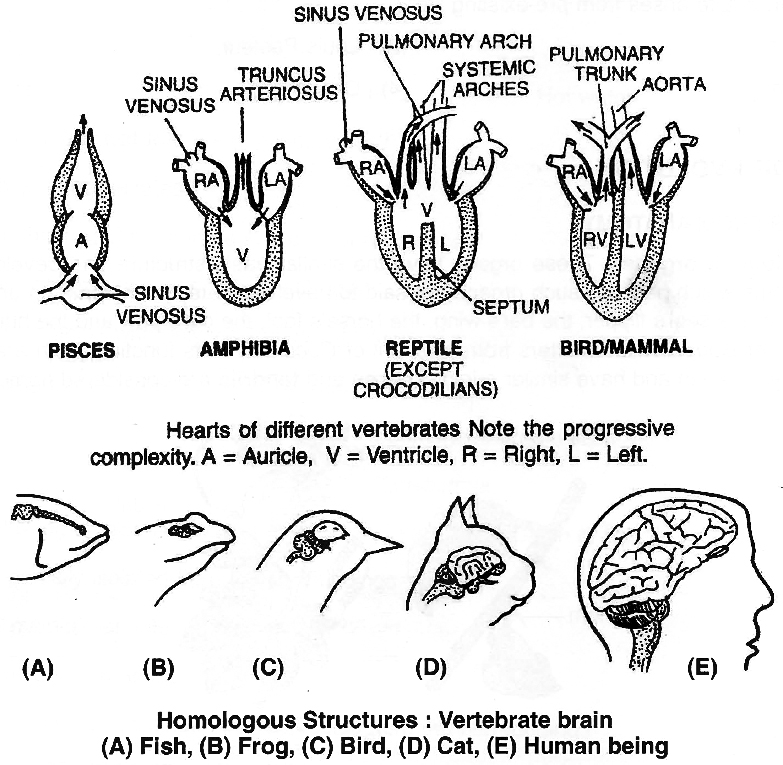
(ii) Analogous organs :
These organs which are not anatomically similar though they perform similar functions.
For example, the wings of birds and of butterfly look alike, they perform the similar function of flying but they are not anatomically or structurally similar.
Even the wings of birds and bats are also analogous structures which have different origins.
Other examples are flippers of penguin and dolphin (the former is a bird and the later is a mammal); eye of an octopus and the eye of a mammal, both differ in retinal position, still the function is same.
In plants, sweet potato (root modification) and potato (stem modification) is another example of analogy.
Both are meant for storage of food but modifications of different parts of plant.
Now, what is the reason of development of analogous structures?
The possible explanation may be that it is the similar habitat that has resulted in selection of similar adaptive features in different (distantly related) groups of organisms put toward the same function.
This phenomenon is termed adaptive convergence or convergent evolution which is the opposite of adaptive radiation as seen in the homologous structures.
(iii) Vestigial organs:
They are believed to be remnants of organs which were complete and functional in their ancestors.
The study of vestigial organs offer an evolutionary explanation of such rudimentary vestiges by stating that adaptations to new environment of the organism have made these structures redundant.
Such structures are called vestigial organs.
The rudiment of the reptilian jaw apparatus, the rudiment of the hind limbs of python and Greenland whales are some of the examples of vestigial organs.
In humans, many vestigial structures indicate a relationship to other mammals, including the primates.
For instance, muscles of the external ear and scalp are rudimentary and often non-functional.
But these are common to many mammals where they are functional.
The reduced tailbones and nictitating membrane of the eye, the appendix of the caecum, rudimentary body hair and wisdom teeth -all are examples of vestigial organs.
The appendix of man is thought to be a remnant of the large caecum -the storage organ for cellulose digestion in herbivorous mammals.
Similarly, the non-functional vestiges of the pelvic girdle in python and porpoise show, for instance, that the snake and the porpoise originally evolved from four footed ancestors.
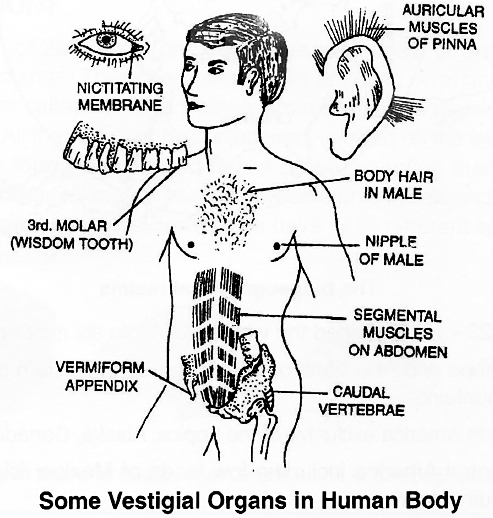
(iv) Atavism:
Sudden reappearance of ancestral character is called atavism.
For example, tail in new born human baby.
The winged petiole of Citrus represents that the unifoliate condition is derived from the trifoliate leaf.
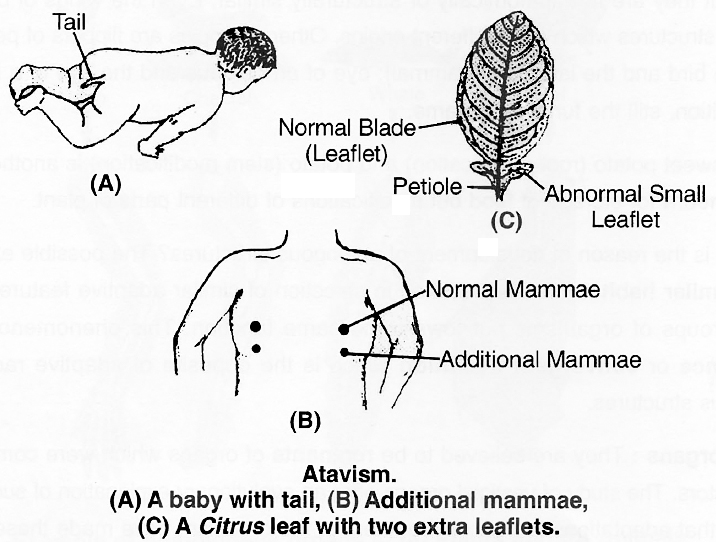
Adaptive radiation
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
WHAT IS ADAPTIVE RADIATION?
Galapagos Islands are chain of 22 islands, present on the west coast of South America.
During his journey, Darwin went to Galapagos Islands.
There he observed an amazing diversity among creatures.
Of particular interest, small black birds, later called Darwin's finches, amazed him.
He realised that there were many varieties of finches in the same island.
All the varieties, he conjectured, evolved on the island itself.
From the original seedeating features, many other forms with altered beaks arose, enabling them to become insectivorous and vegetarian finches.
These birds in Galapagos island show differences in bills and feeding habits, but still resemble with the birds present on original mainland.
Hence, we have seen different species have evolved from single common ancestor.
And this process of evolution of differentm species in a given geographical area starting from a point and literally radiating to other areas of geography (habitats) is called adaptive radiation of which the Darwin's finches represent one of the best example.
Concept Builder
The evolution of Darwins finches on the Galapagos Islands is a classic example of species formation and illustrates how adaptation to local conditions produces the divergence, that is the heart of species formation, i.e., such an evolutionary process, giving rise to new species adapted to new habitats and ways of life, is called Adaptive Radiation or Divergent Evolution.
The clusters of species that have been formed on the Galapagos Islands are thus a Tasmanian wolf clear example of species formation arising by microevolutionary divergence from an ancestral form occupying different habitats of microevolution leading to macroevolution.
Another example is Australian marsupials.
A number of marsupials, each different from the others, evolved from an ancestral stock, but all within the Australian continent.
When more than one adaptive radiation appeared to have occurred in isolated geographical areas (representing different habitats), one can call this convergent evolution.
Placental mammals in Australia also exhibit adaptive radiation in evolving into varieties of such placental mammals each of which appears to be 'similar' to a corresponding marsupial (e.g., Placental wolf and Tasmanian wolf marsupial)
Placental mammals in Australia also exhibit adaptive radiation in evolving into varieties of such placental mammals each of which appears to be 'similar' to a corresponding marsupial (e.g., Placental wolf and Tasmanian wolf marsupial)
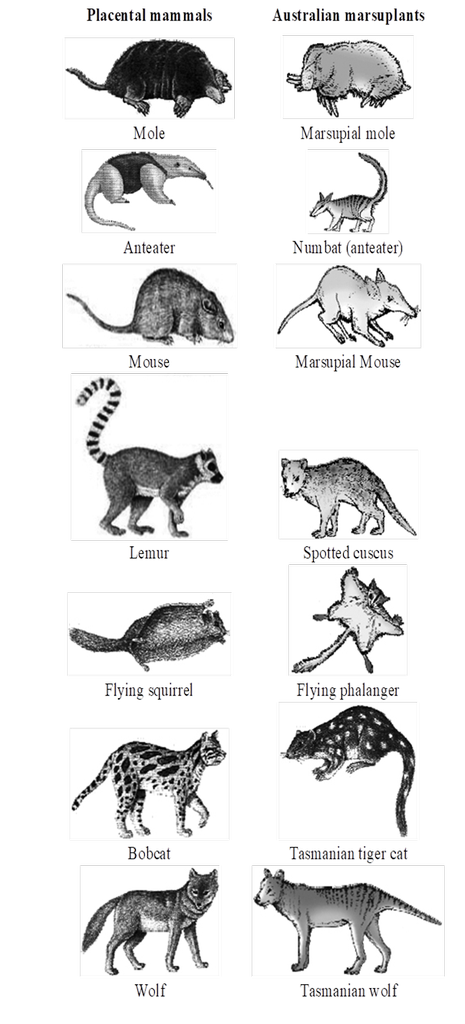
Picture showing convergent evolution of
Australian marsupials and placental mammals
3. EMBRYOLOGICAL EVIDENCES
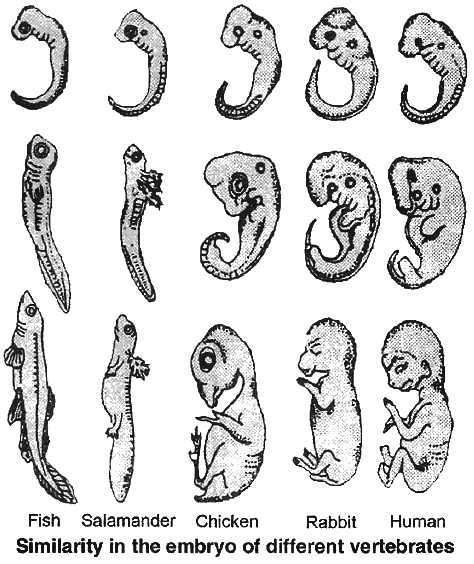
The sequence of embryonic development in different vertebrates show striking similarities.
Gill clefts and notochord appear in the embryonic development of all vertebrates from fishes to mammals.
The notochord is replaced by the vertebral column in all adult vertebrates.
Similarly, gills are replaced by lungs in adult amphibians, reptiles and mammals.
Such similarities in embryonic development once again reinforce the idea of evolution from common ancestors.
Occasionally, embryonic features such as the tail and gill slits persist in adults.
According to Ernst Haeckel, ontogeny (development of embryo) is recapitulation of phylogeny (the ancestral sequence).
This view was summarised by his Biogenetic Law : Ontogeny Recapitulates Phylogeny.
Developmental evidence for evolution is also available from plants.
It is generally believed that mosses and ferns are more evolved than algae.
Protonema of mosses resembles certain green algae.
This provides a clue to their evolutionary relationship.
Both bryophytes and pteridophytes have ciliated sperms and require water for fertilisation.
Gymnosperms do not need water for fertilisation.
But Cycads and Gingko, the primitive gymnosperms, have ciliated sperms like the pteridophytes.
This suggests that gymnosperms have descended from pteridophyte-like ancestors.
The occurrence of ancestral traits in embryo is called Palaeogenesis.
4. PALAEONTOLOGICAL EVIDENCES
Fossils are the remains and/or impressions of organisms that lived in the past last few centuries and palaeontologists have painstakingly built up extensive collections of fossils from all over the world.
The fossil record has helped in building the broad historical sequence of biological evolution.
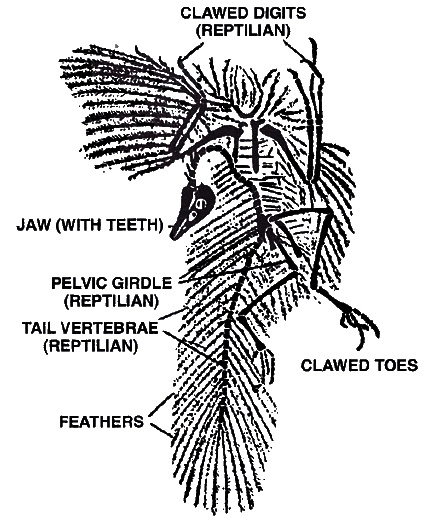
Phylogeny, the evolutionary history of the organism, can sometimes be reconstructed with the help of fossils.
Horse, elephant and man are good examples of relatively complete reconstructions of phylogeny.
Besides form and structure, the habits and behaviour of extinct species can be inferred from the well-preserved fossils.
It is also possible to reconstruct the entire habitat of an organism from fossils.
Fossils also indicate the connecting links between two groups of organisms.
Archaeopteryx shows features of both reptiles and birds.
Reptilian characters of Archaeopteryx.
(a) The body axis is more or less lizard-like
(b) A long tail is present.
(c) The bones are not pneumatic.
(d) The jaws are provided with similar teeth.
(e) Presence of a weak sternum.
(f) Presence of free caudal vertebrae as found in lizards.
(g) The hand bears typical reptilian plan and each finger terminates in a claw.
Avian characters of Archaeopteryx.
(a) Presence of feathers on the body.
(b) The two jaws are modified into a beak.
(c) The fore limbs are modified into wings.
(d) The hind-limbs are built on the typical avian plan.
(e) An intimate fusion of the skull bones as seen in the birds.
By careful analysis of the distribution of fossils in different strata of rocks, the time in history when different species were formed or became extinct can be inferred.
Time Line of Evolution
When scientists first began to study and date fossils, they had to find some way to organise the different time periods from which the fossils came.
They divided the earth's past into large blocks of time called eras.
Eras are further sub-divided into smaller blocks of time called periods, and some periods, in turn, are sub-divided into epochs.
The major geological eras, with their approximate dates in millions of years as given in the table.
"The geological time scale."
Table-I: THE GEOLOGICAL TIME SCALE
Concept Builder
Types of Rocks –
(a) Sedimentary Rocks –
These are formed at the bottom of ancient oceans by deposition of sediments of sand, lime, coal and minerals which slowly change into hard layers.
Sedimentry rocks are also called as stratified rocks.
Eg. Lime Stone, Sand Stone.
Fossils are mostly found in sedimentary rocks.
(b) Igneous rocks –
Such rocks are formed by ancient volcanic deposits which slowly cooled down and hardened as rocks. Fossiles are absent in it. e.g., Granite rocks.
(c) Metamorphic rocks –
These are formed by change in chemical composition of sedimentary rock and igneous rocks or metamorphosis. Such metamorphosis may be caused by pressure heat and physical movements. e.g. Marble Slate rocks. Fossils are also absent in it – due to chemical changes in the fossils are destroyed.
Types of Fossils
(a) Unaltered Fossils :
In this type whole bodies of extinct organisms are found frozen in ice at the polar regions eg. Wooly mammoths (25000 yrs before extinct fossils were found from Siberian region)
(b) Petrified fossils – Most common type of fossil.
Replacement of organic part by mineral deposits is called petrification.
These fossils consists of only the hard parts e.g. bones, teeth, shells, wood etc. of extinct organisms.
In human body first fossilization occurs of teeth.
(c) Mould fossils –
Here no part of the original organism is present, only an impression of the external structure of body is preserved in wet soil.
(d) Cast fossils –
Sometimes minerals fills in the mould, resulting in cast fossils.
(e) Print Fossils –
Foot print or prints of wings, skin, leaves, stems etc made in soft mud which subsequently become fossilized are a common type of fossils.
(f) Coprolites –
These fossils include the fossil preservation of contents of the intestine or excreta of many ancient animals including particularly the reptiles or fishes.
By studying fossils following facts about organic evolution are evident –
1. Fossils found in older rocks are of simple type and those found in newer rocks are of complex types.
2. In the beginning unicellular protozoans were formed from which multicellular animals evolved.
3. Some fossils represents connecting links between two groups
4. Angiosperms among plants and mammals among animals are highly developed and modern organism
5. By fossils, we can study the evolutionary pedigree of an animal like stages in evolution of horse, elephant and man etc.
Fossil Parks
Our country has rich deposits of fossil plants spanning a gap of 3500 million years.
Twenty million years old fossil forests have been discovered and studied by the Birbal Sahni Institute of Palaeobotany, Lucknow. These forests need to be systematically studied and conserved for scientific understanding and enlightenment. Some of the excellent localities that can be raised to the status of national fossil parks are:
1. Fifty million years old fossil forests preserved in the sediments between the streaming lava flow that poured out into the Deccan country at Mandla district, Madhya Pradesh.
2. One hundred million years old fossil forest located in Rajmahal Hills, Bihar.
3. Two hundred and sixty million years old coal-forming forests in Orissa.
Microfossils and Fossil Fuel Exploration
Palaeobiological study helps in understanding and locating coal and hydrocarbon sources.
Palynofossils -tiny microscopic spores, pollen and other vegetal remains of the past -assist us in interpreting ancient environmental conditions favourable for organic matter accumulation and its conversion to fossil fuels by transformation and subsequent thermal alteration.
By quantitative analysis of microfossils, it is possible to determine the approximate location and configuration of near shore marine deposits, which are in turn responsible for formation and accumulation of hydrocarbons.
The main source of hydrocarbons are phytoplankton, marine and terrestrial algae as well as lipid-rich plant remains.
Thus, the study of fossil plants offers an effective tool in stratigraphical geology and can be exploited in tapping organic fuel resources.
Concept Builder
Dating Fossils
Naturally occurring radioactive isotopes of certain elements are employed in this process.
Such isotopes are unstable and decay over the course of time at a steady rate, producing other isotopes.
One of the most widely employed methods of dating -the carbon -14 (14C) method uses estimates of the different isotopes present in samples of carbon.
Most carbon atoms have an atomic weight of 12.
But a fixed proportion of the atoms in a given sample of carbon consists of carbon with an atomic weight of 14 (14C).
14C is produced from 12C as a result of bombardment by particles from space.
But after an organism dies and is no longer incorporating carbon, its 14C gradually decays over time, by the loss of neutrons.
The common radioactive elements which lose their radioactivity and change into their non radioactive isotopes at a fixed rate are :
1. Potassium40 Argon40
2. Carbon14 Nitrogen14
3. Uranium238 Lead207
4. Rubidium87 Strontium87
6. Thorium232 Lead207
The half life of C-14 is 5730 ± 40 years which means in every 5730 ± 40 years, half of C14 will decay back to N14.
Radioactive carbon can be used to determine the age of fossils upto 70,000 years old.
Half life of potassium40 is 1.3 × 109 years.
Potassium -Argon method is useful because potassium is a common element found in all sorts of rocks.
Potassium decays into Argon extremely slowly.
Electron spin resonance method is relatively most accurate method for dating of fossils.
Evolution of Modern Horse
Eohippus (=Hyracotherium)
The evolution of modern horse began in the Eocene epoch.
The first fossil named Eohippus, 'dawn horse', was in North America.
This horse was about the size of a fox or terrier dog (a type of small dog for unearthing foxes), only 40 cm high at the shoulders.
It had short head and neck.
The fore limbs were with four complete fingers (2, 3, 4 and 5) and one splint of first finger and the hind limbs with three functional toes (2, 3 and 4) and one splint of fifth toe.
Splints are non-functional reduced fingers and toes of horse.
Teeth were with incomplete cement.
Molar teeth had no serrations.
Low-crowned molar teeth were adapted to browse soft lush vegetation.
Mesohippus.
Mesohippus, the intermediate horse, evolved from Hyracotherium about three crore years ago during Oligocene epoch.
It was of the size of modern sheep, about 60 cm high at the shoulders.
Fore feet had three fingers and one splint of fifth finger and hind feet possessed three toes, but the middle one was longer than others and supported most of the body weight.
Molar teeth had some serrations.
Merychippus
Merychippus, the ruminating horse, arose from Mesohippus in Miocene epoch about two crore years ago.
It was of the size of small pony, about 100 cm high at the shoulders.
It had a longer neck.
Its fore and hind limbs had three fingers and three toes, the middle finger and toe being longer than others and supported entire body weight.
There was no splint.
Teeth were longer with cement.
Molar teeth had well developed serrations.
Pliohippus
Pliohippus, the Pliocene horse, evolved from Merychippus in Pliocene epoch about one crore years ago.
It was the size of modern pony, about 120 cm high at the shoulders.
Its each fore and hind limbs had one complete finger and one complete toe and two splints hidden beneath the skin.
Pliohippus is, therefore, referred to be the first one toed horse.
The, molar teeth were long with well developed cement and serrations.
Teeth were adapted for eating grass.
Equus
This is the modern horse which arose from Pliohippus in Pleistocene epoch about nine to ten lakh years ago in North America and later spread throughout the world except Australia.
It is about 150 cm high at the shoulders.
It has a long head and a long neck.
Each fore and hind limb of the modern horse has one finger and one toe and two splints.
The crowns of molar teeth are elongated with enameled ridges and are highly suitable for grinding.
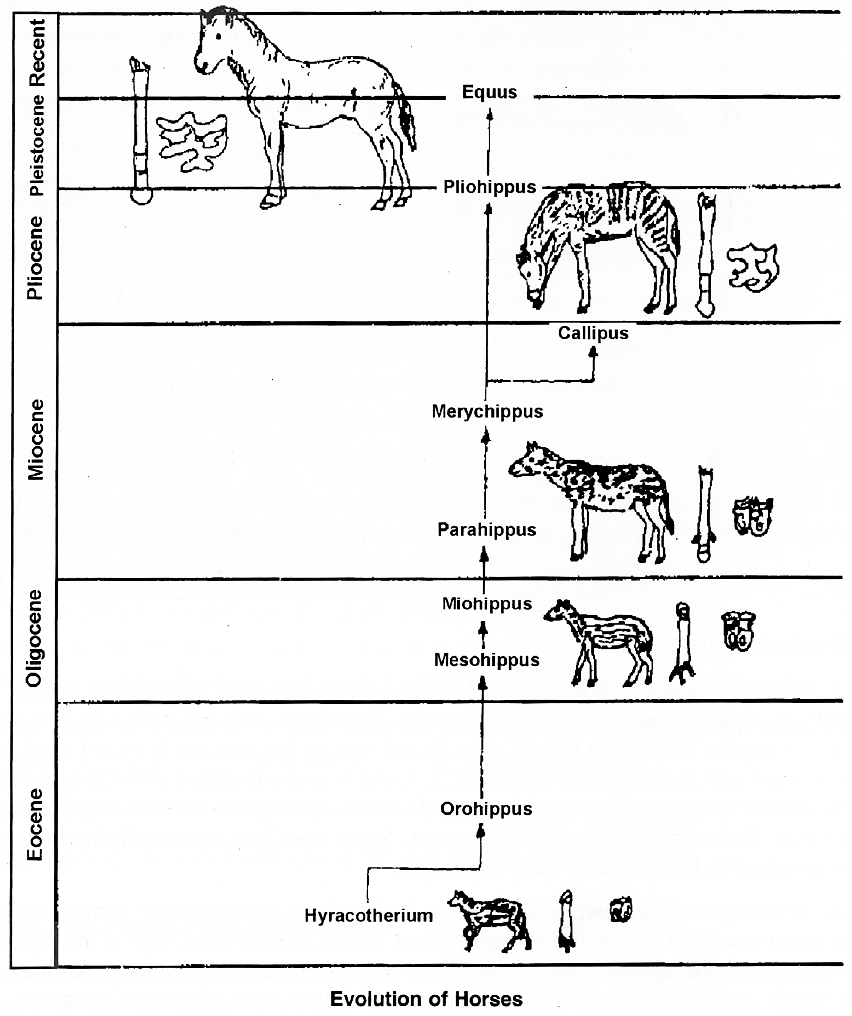
During evolution of horse, there was:
(i) General increase (with occasional decrease) in size,
(ii) Progressive loss of toes,
(iii) Lengthening of toes that was retained,
(iv) Lengthening of limbs in general,
(v) Enlargement of brain, especially cerebral hemispheres,
(vi) Increase in height,
(vii) Increase in the complexity of molar teeth and an enlargement of the last three premolars until they came to resemble molars.
Concept Builder
Connecting Links :
The organisms which possess the characters of two different groups. Examples
1. Proterospongia : A link between Protozoa and Porifera.
2. Neopilina : A connecting link between Annelida and Mollusca.
3. Peripatus : A connecting link between Annelida and Arthropoda.
4. Lungs fishes, e.g., Protopterus, Lepidosiren, Neoceratodus are considered the connecting links between the fishes and amphibians.
5. Egg laying mammals; example duck billed Platypus (Ornithorhynchus) and spiny ant eater (Echidna) are considered connecting links between reptiles and mammals.
Evolution of Vertebrates and Major Groups of Plants
The patterns of evolution of vertebrates and major groups of plants are conspicously different.
The major groups of vascular plants have left relatively small number of fossils which even show gaps (fossilless dark periods).
There are relatively few major lineages, and all the lineages are very distinct from one another.
Instead of showing gradual and continuous change through time, the major lineages appear suddenly in the fossil record.
After that, they persisted with little fundamental change for hundreds of millions of years.
The existence of many of the major subdivisions of the vascular plants living today can be recognised about 345 million years ago on the basis of their distinctive reproductive structure.
All primitive land plants reproduce via tiny spores contained in the sporangia. The major taxonomic groups are distinguished by the position of sporangia on the plant.
The sporangia are terminal, located at the tip of the plant in the most primitive Psilopsida.
These are placed at the base of the leaves in the Lycopsida (represented in the modern flora by Lycopodium and Selaginella).
The sporangia are arranged in whorls at the top of the plant in Sphenopsida (horsetails).
Fossil evidences document that these basic patterns have been maintained for more than 350 million years.
Few, if any, intermediates are known between these patterns.
The origin of seeds in the land plants was achieved about 345 million years ago in lineages recognised as ancestral to all more advanced vascular plants.
The last major evolutionary advancement among the vascular plants was the emergence of flowering plants (the angiosperms) about 140 million years ago.
But the fossils left no clue as to their ancestors.
The fossil records also indicate that nearly all the living orders of angiosperms and most of the characters of their modern-day representatives evolved from them.
The continuous change of a character within an evolving lineage is termed as evolutionary trend.
A lineage is an evolutionary sequence, arranged in linear order from an ancestral group to a descendant group.
The number of trends in any lineage is, therefore, same as the number of characters evolving.
A trend may be progressive (a general increase in size of organs) or retrogressive (a general degeneration and loss of organs).
Biological evolution
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
BIOGEOGRAPHICAL EVIDENCES
The study of patterns of distribution of animals and plants in different parts of the earth is called Biogeography.
Alfred Russel Wallace (1823 -1913) divided the whole world into six major biogeographical regions or realms.
1. Palaearctic : Europe and Asia north of the tropics, north-western corner of Africa, including the Atlas Mountains.
2. Nearctic : North America exclusive of the tropics, Alaska, Canada, United States and Mexico.
3. Neotropical : Central America including low lands of Mexico, islands of the Caribbean and all of South America.
4. Ethiopian Africa (with exception of the Atlas Mountains), Madagascar and adjacent islands.
5. Oriental Tropical part of Asia (including India) south of the Himalaya Mountains and eastward through Sumatra, Java, Borneo and the Philippines.
6. Australian Australia, Tasmania, New Guinea and all islands of the Indonesian archipelago that lie east of Borneo, beginning with Celebes.
Biogeographic map of the world is that in which the six major biogeographic realms are present.
Geologists believe that millions of years ago, all the continents we demarcate today, were in the form of a single land mass.
On account of geological changes, especially movements of crustal plates below the surface of the earth, huge land masses broke off and drifted apart from one another.
As these land masses (continents) moved away, the seas separated them and acted as barriers to the free movement of organisms among the continents.
Because of variable environmental conditions prevailing on the different continents, over centuries, plants and animals evolved independently in each biogeographical region.
Concept Builder
India falls under Oriental realm. Two geographical regions separated by a high mountain ranges are Palaearctic and Oriental.
Consider, for example, two instances which show similarities in the pattern of distribution of plants and animals between two land masses which were once part of a larger land mass.
1. The flora and fauna on each of the Galapagos Island -a chain of 22 islands in the Pacific ocean on the west coast of South America resemble those of the South American mainland with which the Galapagos Islands were once connected.
2. Magnolias, Tulips and Sassafras are found naturally growing in the eastern USA and in China. Hence, these show disjunct distribution which means that these flora have different groups that are related but widely separated geographically.
The distribution pattern of the present-day animals and plants as well as the distribution of fossils are best explained on the basis of the theory of evolution.
The birds in Galapagos Islands show differences in bills and feeding habits.
The bills of several of these species resemble those of different, distinct families of birds on the mainland.
All these birds are thought to have evolved from a single common ancestor.
Mechanism of evolution?
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
Mechanism of Evolution
Populations evolve, but individual organisms do not. A population is an interbreeding group of individuals of one species in a given geographic area at the same time. A population evolves because the population contains the collection of genes called the gene pool. As changes in the gene pool occur, a population evolves.
- Hugo de Vries worked on evening primrose. He gave the idea of mutations.
- Mutation is the difference arising suddenly in a population. Hugo de Vries theory of mutation differs from Darwin
- He stated that mutation causes evolution and not the minor variations that as suggested by Darwin.
- Mutations are sudden, random and directionless while Darwinian variations are small and directional.
- Evolution according to Darwin was slow and gradual whereas Hugo de Vries believed that mutation caused large changes that led to speciation. He therefore called it is saltation (single step large mutation).
- Saltation: It is a large and an abrupt evolutionary change that has been brought about due to sudden large-scale mutation.
Hardy weinberg principle
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
HARDY -WEINBERG PRINCIPLE
Five basic processes affect the Hardy Weinberg equilibrium and cause variations at genetic level. These are:
(i) Mutation
(ii) Gene migration
(iii) Genetic drift
(iv) Recombination
(v) Natural selection
The Hardy-Weinberg principle states that the proportions of different alleles will stay the same in a large population if mating occurs at random and the above mentioned forces are absent.
In algebraic terms, the Hardy-Weinberg principle is written as an equation.
Its form is what is known as a binomial expansion.
For a gene with two alternative alleles, called A and a, the frequency of allele A can be expressed as p and that of alternative allele a as q, because these are only two alleles, p + q must always be equal to one. The equation looks like this
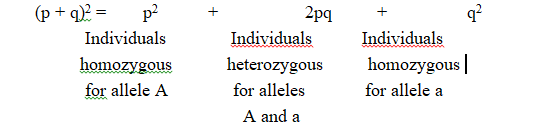
For example, if q is the frequency of the allele a, then the Hardy-Weinberg equation states that q2 = percentage individuals homozygous for allele a say 16%.
q2 = 0.16, q = 0.4
Concept Builder
Factors of Evolutionary Change
(i) Mutation (ii) Migration (iii) Genetic Drift
(iv) Recombination (v) Natural Selection
(I) MUTATION
Replica Plate Experiment of Lederberg and Lederberg
1. Mutations are random (indiscriminate) with respect to the adaptive needs of organisms.
2. Most mutations are harmful or with no effect (neutral) on their bearer.
3. Mutation rates are very slow.
The Lederberg Replica Plating Experiment, a beautiful example of the genetic basis of a particular adaptation was demonstrated in bacteria by an ingeneous method devised by Joshua Lederberg and Esther Lederberg.
E.coli bacteria are usually grown in the laboratory by plating dilute suspensions of bacterial cells on semi-solid agar plates.
After a period of growth, discrete colonies appear on the agar plates.
Each of these colonies originates from a single bacterium through a large number of cell divisions.
The Lederbergs inoculated bacteria on an agar plate and obtained a 'master plate' containing several bacterial colonies.
They, then created several replicas of this master plate by a simple procedure.
A sterile velvet disc, mounted on a wooden block, was gently pressed on the master plate.
Some bacteria from each colony adhered to the velvet.
By pressing this velvet on to new agar plates, they obtained exact replicas of the master plate, because the few bacteria transferred by the velvet formed colonies on the new agar plates.
However, when they attempted to make replicas using plates containing an antibiotic such as penicillin, most colonies found on the master plate did not grow on the replica plates.
The few colonies that did grow were obviously resistant to penicillin.
How did the bacteria acquire the ability to grow in a new environment (here, agar medium, containing penicillin)? In other words, what was the origin of this adaptation?
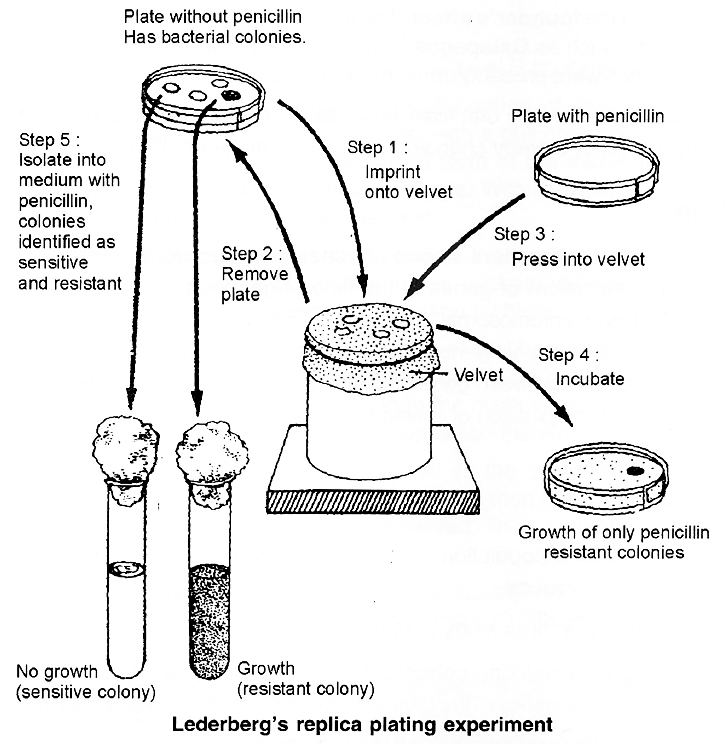
A Lamarckian interpretation of this adaptation would have been that penicillin somehow induced a change in one or more bacteria, enabling them to grow in the presence of penicillin.
A Darwinian view is that there were, in the original suspension of bacteria from which the master plates were prepared, a few bacteria carrying mutant genes which conferred on them the ability to survive the action of penicillin and form colonies.
These mutations, which had arisen by chance, and not induced by penicillin, were present only is small numbers in the original suspension.
Lederberg's experiment provided evidence that mutations are actually preadaptive.
These kinds of mutations are regarded as advantageous mutations.
They appear without exposure to the environment.
Actually, the preadaptive mutations express themselves only after exposure to the new environment to which the organisms are to adapt themselves.
The new environment does not induce the formation, it only selects the preadaptive mutations that occurred earlier.
(II) MIGRATION
Migration, defined in genetic terms as the movement of individuals from one population into another, can be a powerful force in upsetting the genetic stability of natural populations.
If the characteristics of the newly arrived animal differ from those already there, the genetic composition of the receiving population may be altered, if the newly arrived individual or individuals can adapt to survive in the new area and mate successfully.
Gene pool : A total collection of all genes and its allele in a population is called gene pool. Thus, gene pool will have all genotypes i.e., genes of the organisms.
Gene flow: If genes are exchanged between two different populations of a species, it is gene flow.
(III) GENETIC DRIFT I SEWALL WRIGHT EFFECT I NON-DIRECTIONAL FACTOR
Natural selection is not the only force responsible to bring about changes in gene frequencies. There is the role of chance or Genetic Drift also.
Genetic Drift causes the change in gene frequency by chance in a small population.
In a small population, the one individual alleles of a gene are represented by a few individuals in a population.
These alleles will be lost if these the individuals fail to reproduce.
Allele frequencies appear to change randomly, as if the frequencies were drifting, Janes thus, a random loss of alleles in small population is Genetic Drift.
A series of small populations that are isolated from one another may come to differ strongly as a result of Genetic Drift.
Genetic Drift has two ramifications are described below.
1. Bottle neck effect:
It is the decrease in genetic variability in a population, e.g., cheetah population in Africa decreased due to hunting.
Their decreased numbers have limited cheetahs genetic variability, with serious consequences.
The present cheetah population is susceptible to a number of fatal diseases.
If any of these diseases attacks the cheetah population, the path of extinction of cheetah cannot be reversed.
2. Founder's effect:
When one or a few individuals are dispersed and become the founders of a new, isolated population at some distance from their place of origin, the alleles that they carry are of special significance.
Even if these alleles are rare in the source population, they will be a significant fraction of the new population's genetic endowment.
This effect by which rare alleles and combinations of alleles may be enhanced in new populations -is called the founder's effect.
The founders effect is particularly important in the evolution of organisms on islands, such as Galapagos Islands which Darwin visited.
Most of the kinds of organisms that occur in such areas were probably derived from one or a few initial founders.
Fixation of new mutations:
Genetic drift fixes new alleles, genes that arise by mutation, from time to time and eliminate the original gene, thereby changing the genetic make up of small population.
(IV) RECOMBINATION
Gene recombination is also an important source of variations.
It occurs during crossing over at the time of meiosis, free assortment (selection) of genes at the time of gamete formation, random union of gametes at the time of fertilization and even chromosomal aberrations.
They cause reshuffling of gene recombinations which provide new combinations of existing genes and alleles.
This is the entity of gene recombination.
Gene recombination can occur not only between genes but also within genes resulting in the formation of a new allele.
Since it adds new alleles and combination of alleles to the gene pool, it is an important process during evolution which causes variations.
(V) NATURAL SELECTION
It causes allele frequencies of a population to change. Depending upon which traits are favoured, natural selection can produce different results.
Forms of Selection: There are three kinds of natural selections
Stabilizing Selection (Normalizing selection) :
When selection acts to eliminate both extremes from an array of phenotypes, the frequency of the intermediate type which is already the most common, is increased.
Directional Selection (Progressive selection) :
When selection acts to eliminate one extreme from an array of phenotypes, the genes determining this extreme become less frequent in the population. The industrial melanism is peppered moth, Biston betularia provides good example of directional selection from nature.
Disruptive Selection (Diversifying selection) :
In some situations, selection acts to eliminate, rather than favour, the intermediate type. The individuals at both the extremes are favoured.
Examples of Natural Selection-Industrial Melanism
First studied by R.A. Fisher and E.B. Ford and in recent time by H.B.D. Kettlewell.
One of the most striking examples, which demonstrates the action of natural selection, is the industrial melanism in England.
The peppered moth Biston betularia, with a dull grey colour or white was abundant in England before the Industrial Revolution.
A black coloured form of the same moth (melanic, a dominant mutant differing in a single gene), carbonaria, was very rare.
Within a couple of hundred years, however, the proportion of carbona ria increased to almost 90 per cent.
The moths rest on tree trunks.
Before the Industrial Revolution, the tree trunks used to be covered with grey coloured lichen.
The dull grey moth easily blended with this background, while the black moth stood out conspicuously, and was therefore more susceptible to predation by birds.
With the advent of the Industrial Revolution, large-scale burning of coal became common.
The enormous amount of smoke produced resulted in the deposition of particulate matter on tree trunks, turning them black.
As a result, the grey moths now became more conspicuous than the black variety, and hence more susceptible to predation.
The frequency of black coloured moths in the population therefore increased.
Gradual replacement of coal by oil and electricity, as well as the improved methods of controlling soot production, reduced the soot desposition on the trees.
Conditions then became more suitable for the survival of grey moths, consequently their frequency once again increased.
Thus, reduction in pollution is now correlated with reverse evolution.
Industrial melanism, as this phenomenon is called, is thus a particularly interesting example which clearly brings out the action of natural selection.
This has been observed in about 70 different species of moths, and in several other European countries as well.
This understanding is supported by the fact that in areas where industrialisation did not occur e.g., in rural areas, the count of melanic moths was low.
This showed that in a mixed population, those that can better-adapt, survive and increase in population size.
Remember that no variant is completely wiped out.
Similarly, excess use of herbicides, pesticides, etc., has only resulted in selection of resistant varieties in a much lesser time scale.
This is also true for microbes against which we employ antibiotics or drugs against eukaryotic organisms/cell.
Hence, resistant organisms/cells are appearing in a time scale of months or years and not centuries.
These are examples of evolution by anthropogenic action.
This also tells us that evolution is not a direct process in the sense of determinism.
It is a stochastic process based on chance events in nature and chance mutation in the organisms.
Change In Genotypic Frequencies
If the alleles for grey and black colours are denoted by G and B, the genotypes of the moths would be GG, GB and BB.
Since B is dominant, GB and BB will be black.
Due to greater predation by birds on the black (melanic) phenotype the proportion of B in the population was maintained at a much lower value than G.
Resistance of Mosquitoes to Pesticides
Mosquitoes have always been a major health hazard, especially as they are responsible for the spread of diseases such as malaria and filaria.
When DDT was first introduced to control mosquitoes, it was tremendously successful; most mosquitoes were sensitive to DDT and were therefore killed.
However, DDT has now become ineffective against mosquitoes.
This is explained as follows:
In the original population of mosquitoes, some individuals were resistant to DDT.
However, in the absence of DDT, such resistant individuals were few because they had no advantage over the DDT-sensitive mosquitoes.
However, when DDT was used on a large-scale, only the resistant genotypes were able to survive and reproduce.
As a result, over a period of time, almost the entire population came to consist of the resistant type, which made DDT quite ineffective.
Evolution is thus a change in gene frequencies in the population in response to changes in the environment-in this case the introduction of DDT.
The principle of natural selection thus helps us to understand, why such chemical insecticides would remain useful only for a limited time.
Concept Builder
Plants Growing Around Mines
A few plants are now known to grow on the tailings or refuse around mines. Professor A.D. Bradshaw studied one such grass, the bent grass Agrostis tenuis growing on tailings of lead mines in Wales, U.K.
He took some of these and planted them in soil from a pasture nearby.
Similarly, he transplanted live Agrostis plants from the pasture to the lead-rich soil.
The bent grass from the mine soil grew very slowly on normal pasture soil.
The one from the pasture, on the other hand, could not survive in the lead-rich soil.
A very small percentage (three out of sixty), however, could grow in the soil rich in lead.
These were undoubtedly the kind, from which the race of bent grass capable of growing in lead-rich mine soil evolved originally.
Plants tolerant to selenium such as Astragalus and Haplopappus have been reported from the U.S.A.
These plants are not only capable of growing in seleniferous soils, but require selenium as an essential element.
In our country, Professor Y.D. Tyagi discovered populations of Impatiens balsamina growing around Zwar zinc mines in Udaipur, Rajasthan.
The presence of such plants, which have evolved metal tolerance, can indicate the occurrence of specific metal deposits.
Such plants are called bioindicator plants.
Sickle Cell Anaemia Is an example of balancing selection.
(i) In few RBCs, 1-2% became sickle shaped during lack of oxygen.
(ii) The heterozygotes (HbA / HbS), who have one copy of sickle cell allele, coupled with one normal allele are better survivors in the areas where malaria is endemic; because the malarial parasite spends a part of the life cycle in the RBC; if they enter into the RBC which are sickle shaped, they will die.
(iii) The women who are heterozygote have higher fertility; that's why natural selection has not eliminated the allele.
(iv) The loss of deleterious recessive genes through deaths of homozygotes (HbS / HbS) is being balanced by the gain resulting from successful reproduction by heterozygotes in malaria prone areas. For this reason, the selection is called balancing selection.
(v) Heterozygotes enjoy some resistance to malaria, so they survive the malarial parasite more successfully than either normal or sickle cell homozygotes.
ARTIFICIAL SELECTION
Some genetic variability is always present in a population.
Some alleles make organisms better adapted to the environment, and thus make them more successful in survival and reproduction.
As a result, the frequency of such alleles in a population gradually increases.
This is called selection; these alleles are thus 'selected' over the other alleles.
This process operating in natural populations is therefore called 'Natural Selection'.
The process of natural selection, acting on variability inherent in the population, over millions of years, has given rise to the great diversity we see in the biological world.
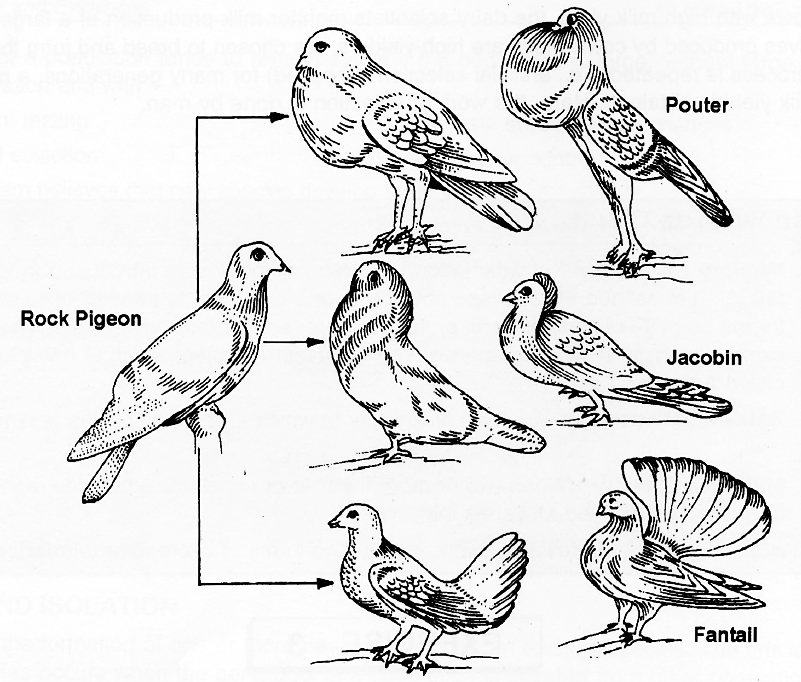
Ancestry of different breeds can be traced to wild rock pigeon. (Artificial Selection)
Man has been using a similar process for improving the qualities of domesticated plants and animals for centuries.
Plant-breeding and animal-breeding are very similar to the action of natural selection, the difference being that the role of nature is played by man.
The criteria for selection are based on human interests.
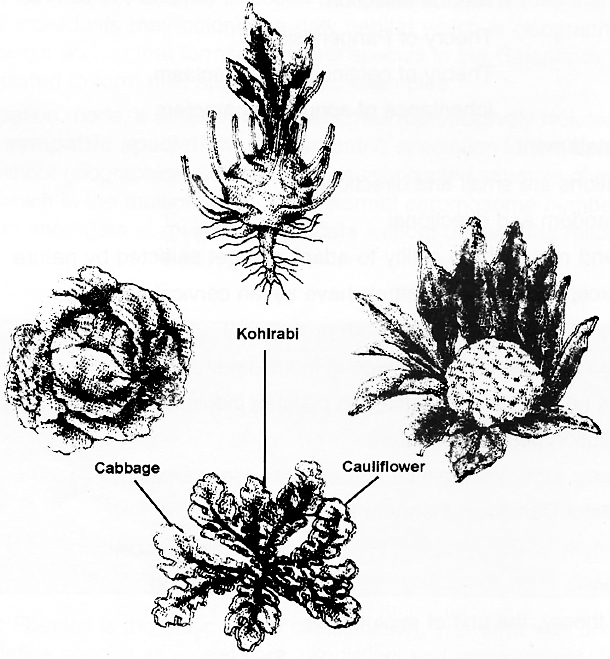
of a common ancestor, colewort (Artificial Selection)
To obtain cows with high milk yield, the dairy scientists monitor milk production of a large number of cows.
Only the calves produced by cows which are high-yielders, are chosen to breed and form the next generation.
When this process is repeated (i.e., artificial selection is applied) for many generations, a population of cows with high milk yield is obtained.
Here, the work of selection is done by man.
Concept Builder
Mimicry: It is a kind of adaptation. The term mimicry was introduced in Biology by Bates (1862). It is defined as "the resemblance of one organism to another or to any natural object for the purpose of concealment, protection or for some other advantages. The organism which exhibits mimicry is called a mimic. The organism or object which is mimicked or imitated is called a model.
Bateslan Mimicry : It is a form of mimicry in which an edible species resembles an inedible one.
Mullerian Mimicry : When two or more inedible or unpalatable species resemble each other the mimicry is termed Mullerian mimicry.
Both Batesian and Mullerian mimicries are two forms of protective mimicries.
Speciation and isolation
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
SPECIATION AND ISOLATION
Speciation is the formation of one or more new species from an existing species.
The crucial episode in the origin of species occurs when the gene pool of a population is severed from other populations of the parent species and gene flow no longer occurs.
Speciation can take place in two modes based on the geographical relationship of a new species to its ancestral species.
1. When a population, formerly continuous in range, splits into two or more geographically isolated populations and form new species, the mode of speciation is called allopatric speciation.
This can happen by subdivision of the original population, when a geographical barrier, such as a creeping glacier, a land bridge (e.g., Isthmus of Panama) or ocean or mountain, cuts across a species range.
Alternatively, a small number of individuals may colonise a new habitat which is geographically separated from the original range.
Darwin's finches that formed separate species in the Galapagos islands and the Australian marsupials that radiated to form new species are its examples.
2. In the second speciation mode, a subpopulation becomes reproductively isolated in the midst of its parent population; this is sympatric speciation.
So, sympatric speciation is the formation of species within a single population without geographical isolation.
The usually quoted example of sympatric speciation comes from polyploidy, which is the multiplication of the normal chromosome number.
This can happen when chromosomes fail to segregate at meiosis or replicate without undergoing mitosis.
Concept Builder
(I) Multiplicative speciation (Cladogenesis) : It is the formation of two or more species from a single species, example, allopatric speciation, sympatric speciation. This can be gradual or abrupt.
(II) Fusion species : It is an allogenous transformation. Isolation mechanism may break down due to a mutation. The two species will interbreed and merge to form a single species.
(III) Phyletic speciation (Anagenesis) : It is autogenous transformation of a species with passage of time due to piling up of variations.
3. Species concept:
Species is the basic unit of classification.
The term was coined by John Ray (1693).
Most taxonomists define species as morphologically distinct and reproductively isolated natural population or group of populations where individuals resemble one another more closely than with members of other species, have a similar anatomy, karyotype and biochemicals, interbreed freely and form a genetically closed system. There are three basic concepts about the species.
4. Morphospecies concept:
It is the earliest concept of species.
Davis and Heywood (1963) have defined it as "assemblage of individuals with morphological features in common and separable from other such assemblages by correlated morphological discontinuities in a number of features."
However, the number of morphological characters chosen for separating species varies from taxonomist to taxonomist.
"Lumpers" will combine all the populations with broadly similar traits into a single species while "Splitters" will separate various populations with even minor morphological differences into distinct species.
5. Biological species concept:
Though first proposed by Buffon (1753), biological species concept was formulated by Mayr(1942).
According to it, a biospecies (biological species, Biological Species Concept) is sexually interbreeding or potentially interbreeding group of individuals which is reproductively isolated from other species and is therefore, separated from others by absence of genetic exchange.
Normally species are distinct from one another by both morphological traits and reproductive isolation.
However, sibling species are those distinct species which are almost identical morphologically but are distinct from each other due to absence of interbreeding, e.g., Drosophila pseudoobscura and D. persimilis.
Biological species concept is, therefore, mainly based on absence of cross fertilisation between members of two species.
Cross fertilisation tests carried out by taxonomists between individuals of morphological and geographically separated populations have, resulted in revision of species and grouping of many of them into single species, e.g., several species of North American sparrows as subspecies and races of a single song sparrow, Passarella melodia.
The only problem of using reproductive isolation is the absence of sexual reproduction in several organisms -prokaryotes, some protists, some fungi, some plants (e.g., commercial Banana) and animals.
Further, cross fertilisation experiments cannot be performed on such a large number of species that occur in varied geographical areas.
Reproductive isolation cannot be used as a criterion in case of fossils.
The living organisms and fossils can be grouped only on the basis of their morphology and biochemistry.
Mayr (1987) has named morphologically grouped asexual species as paraspecies while Ghiselin (1987) has named them pseudospecies.
6. Evolutionary species concept:
All evolutionary taxonomist have been in search of a proper definition of species which is basic unit of classification.
One such definition has been given by Simpson.
According to Simpson (1961) "an evolutionary species is a lineage (an ancestor-descendent sequence of population) evolving separately from others and with its own unitary evolutionary role and tendencies."
The concept stresses on evolutionary isolation with sexual isolation being its one aspect.
It is more dependent on differences which can be morphological, genetical, behavioural and ecological, to know evolutionary distance.
However, evolution does not occur simultaneously in all the traits.
Neither its rate nor direction (in which it is occurring) are the same.
7. Reproductive isolation may be defined as the existence of intrinsic barrier to the interbreeding in natural populations. Each of these intrinsic barriers is called a reproductive isolating mechanism. According to Mayr (1942), reproductive isolating mechanisms are the biological properties of individuals which prevent the interbreeding of naturally sympatric populations.
8. Reproductive isolation in the form of hybrid sterility is known since long. In the laboratory or in zoos, hybrids can be produced between species that do not interbreed in nature. Horses and donkeys are two different species; a hybrid, mule, is produced from the mating of a male donkey and a mare (female horse).
9. Similarly, mating between stallion (male horse) and female donkey results in a hybrid called hinny. Both mule and hinny are sterile.
10. There are examples of species, which can produce fertile hybrids in captivity. You might have heard about the famous 'tigons', a hybrid of African lioness (Panthera leo) and Asian tigers (Panthera tigris), which is fertile. No barrier to hybridisation between these species has evolved during their long isolation from each other Natural selection has not favoured a reduction in hybridisation for the simple reason that no hybridisation has been possible. Other examples of species that breed in captivity and produce fertile hybrids are mallard (a duck) and the pintail duck, the polar bear and the Alaskan brown bear and the platy and swordtail fishes. But these species do not interbreed at all in natural condition.
Concept Builder
BARRIERS TO HYBRIDISATION
Prezygotic Mechanisms: (Prevent mating or formation of zygote)
1. Ecological isolation : Two species live in different habitats and do not meet. (One may be living in fresh water and the other in the sea).
2. Temporal isolation : Breeding seasons or flowering time may be different in the two species.
3. Behavioural isolation: The males of one animal species are unable to recognise the females of another species as potential mates.
4. Mechanical isolation: The structural differences in genitalia of individuals belonging to different animal species interfere with mating.
5. Gametic isolation : The sperms and ova of different species of animals are unable to fuse. In plants, the pollen coming from a different species may be rejected by the stigma.
Postzygotic Mechanisms : A hybrid zygote is formed but it may not develop into a viable fertile adult.
1. Hybrid Inviability : Hybrid zygotes fail to develop. In plants, embryos arising from interspecific crosses abort.
2. Hybrid sterility : Hybrid adults do not produce functional gametes. (Mules and hinny are common examples in mammals. Several hybrid ornamental plants are sterile.)
3. Hybrid breakdown : The offspring of hybrids are inviable or infertile.
MODERN SYNTHETIC THEORY OF EVOLUTION
1. Evolution on the grand scale of geological time is called macroevolution.
2. Evolution at genetic level is called microevolution.
3. Studies of how individual traits evolve within natural populations provide powerful evidence that natural selection can be a powerful agent of microevolutionary change within species. The progressive change in allele frequencies within the population is micro-evolution.
4. Units of evolution is population.
5. Unit of natural selection is individual.
The modern synthetic theory of evolution is the result of the work of a number of scientists as T. Dobzhansky, R.A. Fisher, J.B.S. Haldane, Sewall Wright, Stebbins.
The synthetic theory includes the following factors.
(i) Gene mutations
(ii) Changes in chromosome structure and number
(iii) Genetic recombination
(iv) Natural selection
(v) Reproductive isolation
(vi) Migration lied
(vii) Hybridisation
NEUTRAL THEORY OF EVOLUTION
According to Kimura most of the mutations are neutral, and are not eliminated from the population.
This is against natural selection.
Kimura proposed that speciation is not due to selection of advantageous genotypes no but elimination of deleterious alleles and random selection of neutral alleles.
It emphasized that most mutations are of neutral value and genetic drift is responsible for divergence.
It means that all mutations are alike in adaptive value.
It is only chance or random drift which delineates a novel collection of mutants into a group divergent from the parental population.
Place of Humans in the Animal Kingdom
Today human evolution is being studied by:
(i) Homology in the chromosomes of man and great apes. The banding pattern of human chromosome number 3 and 6 are compared with those of particular autosomes in the chimpanzee. It shows a common origin for man and chimpanzee.
(ii) Today, besides the autosomal chromosomes, V-chromosomes and mitochondrial DNA are being studied, as they are uniparental in origin and do not take part in recombination.
(iii) Evidence from blood proteins -It has been proved by the blood protein tests that man is most closely related to great apes (chimpanzee and gorilla).
(iv) Evidence from blood groups -The blood groups A and B are found in apes and not in monkeys.
(v) Evidence from haemoglobin -There is 99 percent homology in haemoglobin of man and gorilla.
Human beings are vertebrates and belong to the class mammalia.
Mammals evolved from primitive reptiles in early Triassic period, about 210 million years ago.
But for nearly 150 million years, mammals existed as relatively inconspicuous group of small rat-like creatures, completely dominated by the large number of gigantic reptiles of the Mesozoic age.
It is only after the great extinction of dinosaurs and other large reptiles, that mammals diversified and began to occupy the earth's many different habitats.
Within the class mammalia, human beings belong to the order primates, a group that originated about 65 million years ago and includes not only monkeys and apes but also the lorises, lemurs and tarsiers.
The Anthropoid apes or the ancestors of monkeys, apes and humans evolved about 36 million years ago and the hominids or the ancestors of apes and humans evolved about 24 million years ago.
Today, the apes are represented by two families, namely, Pongidae which include chimpanzees, gorillas and orangutans and Hylobatidae which includes gibbons.
The chimpanzee and gorilla are restricted to Africa, whereas the orangutans and gibbons are found only in Asia.
Humans belong to the family Hominidae in which Homo sapiens is the only living species.
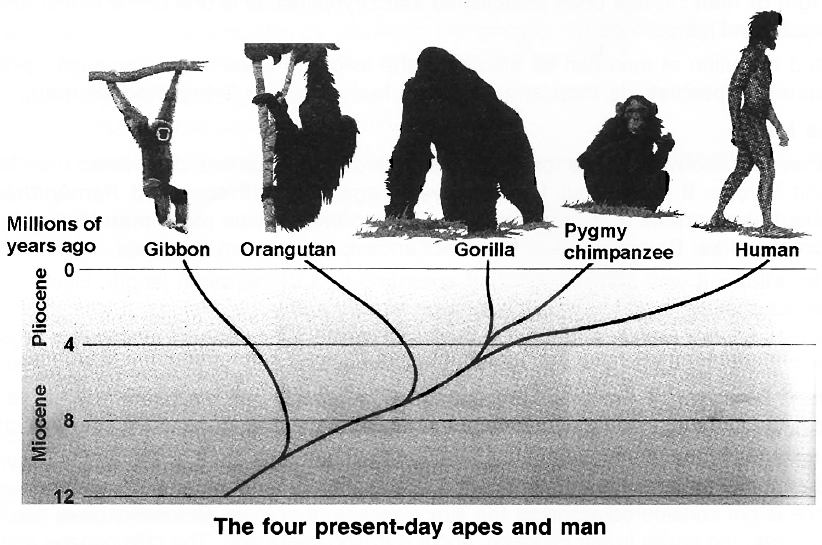
Early Human Ancestors
Tracing the evolution of human beings, both by fossil hunting and molecular methods, is one of the most exciting and active areas of research in biology.
The fossil evidence clearly indicated that such genera as Ramapithecus and Sivapithecus were the forerunners of Hominids.
A genus called Australopithecus appeared in Africa about five million years ago and ultimately gave rise to Homo about two million years ago.
But even Australopithecus had a brain measuring only about 350-450 cm3.
The most important change that must have occurred during the three million years or so, between the appearance of Australopithecus and that of our genus, must therefore have been a phenomenal increase in brain size, all the way up to 1400 to 1450 cm3, that is characteristic of our species.
A combination of molecular data and a modern interpretation of the fossil record suggests that the gibbons probably diverged from the main line of Hominoid evolution about ten million years ago, that the orangutan did so about eight million years ago and that the ancestors of gorilla and chimpanzee, about four million years ago.
The gorilla and chimpanzee have become separated from each other only 2.3 million years ago.
Place and Sequence of Human Evolution
There is evidence that almost all of Hominid evolution occurred in Africa, and Asia and that the evolution of the human species took place in Africa.
Several species belonging to the genus Homo can be recognised from the fossil record. For example, Homo habilis lived in Africa, about two million years ago and was characterised by having a larger brain than Australopithecus, used tools and was bipedal.
Another species, Homo erectus appeared about 1.7 million years ago, used fire and is believed to have migrated to Asia and Europe.
Fossils of the socalled 'Java man' and 'Peking man', belong to Homo erectus. Homo erectus was replaced by Homo sapiens.
A primitive form of Homo sapiens, called Neanderthal man (Homo sapiens neanderthalensis), was common in Europe and Asia.
The Neanderthal men resembled us, though they were relatively short and stocky and more powerfully built.
The Neanderthals made tools and used animal hides as clothing.
They built hut-like structures for dwelling and buried their dead.
There is evidence that an abrupt transition occurred all over Europe whereby the Neanderthal man was wiped out and gave way to the more efficient cousin, the cro-Magnon, about 34,000 years ago.
The Cro-Magnon people left behind very elaborate cave paintings showing the attainment of a form of culture not unlike our own.
After the last glacial period (about 10,000 years ago), modern Homo sapiens sapiens began to spread all over the globe, cultivated plants, domesticated animals and reached enormous population sizes.
Homo sapiens appeared in Africa about 500,000 years and probably replaced Homo erectus there.
But in Asia, Homo erectus appears to have survived for another 250,000 years when it was finally replaced by Homo sapiens migrating from Africa.
Brief of account of evolution
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
BRIEF ACCOUNT OF EVOLUTION
About 2000 million years ago (mya) the first cellular forms of life appeared on earth.
The mechanism of how non-cellular aggregates of giant macromolecules could evolve into cells with membranous envelop is not known.
Some of these cells had the ability to release O2.
The reaction could have been similar to the light reaction in photosynthesis where water is split with the help of solar energy captured and channelised by appropriate light harvesting pigments.
Slowly, single-celled organisms became multi-cellular life forms.
By the time of 500 mya, invertebrates were formed and active.
Jawless fishes probably evolved around 350 mya.
Sea weeds and few plants existed probably around 320 mya.
We are told that the first organisms that invaded land were plants.
They were widespread on land when animals invaded land.
Fish with stout and strong fins could move on land and go back to water.
There are no specimens of these left with us.
However, these were ancestors of modern day frogs and salamanders.
The amphibians evolved into reptiles.
They lay thickshelled eggs which do not dry up in sun unlike those of amphibians.
Again we only see their modern day descendents, the turtles, tortoises and crocodiles.
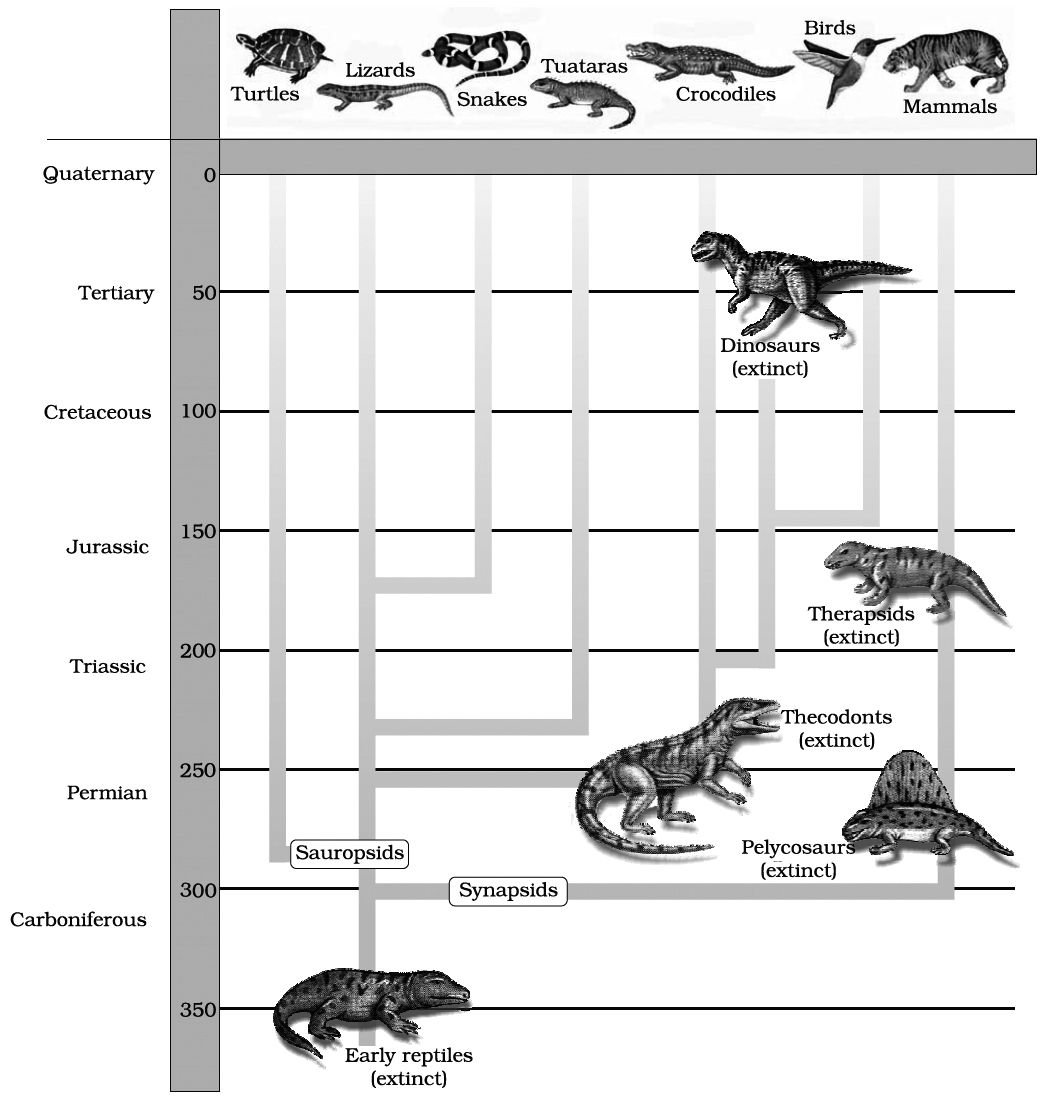
Representative evolutionary history of vertebrates through geological period
This was about 350 mya.
In 1938, a fish caught in South Africa happened to be a Coelacanth which was earlier thought to be extinct.
These animals called lobefins evolved into the first amphibians that lived on both land and water.
In the next 200 millions years or so, reptiles of different shapes and sizes dominated on earth.
Giant ferns (pteridophytes) were present but they all fell to form coal deposits slowly.
Some of these land reptiles went back into water to evolve into fish like reptiles probably 200 my a (e.g. Ichthyosaurs).
The land reptiles were, of course, the dinosaurs.
The biggest of them, i.e., Tyrannosaurus rex was about 20 feet in height and had huge fearsome dagger like teeth.
About 65 mya, the dinosaurs suddenly disappeared from the earth.
We, do not know the true reason. Some say climatic changes killed them.
Some say most of them evolved into birds.
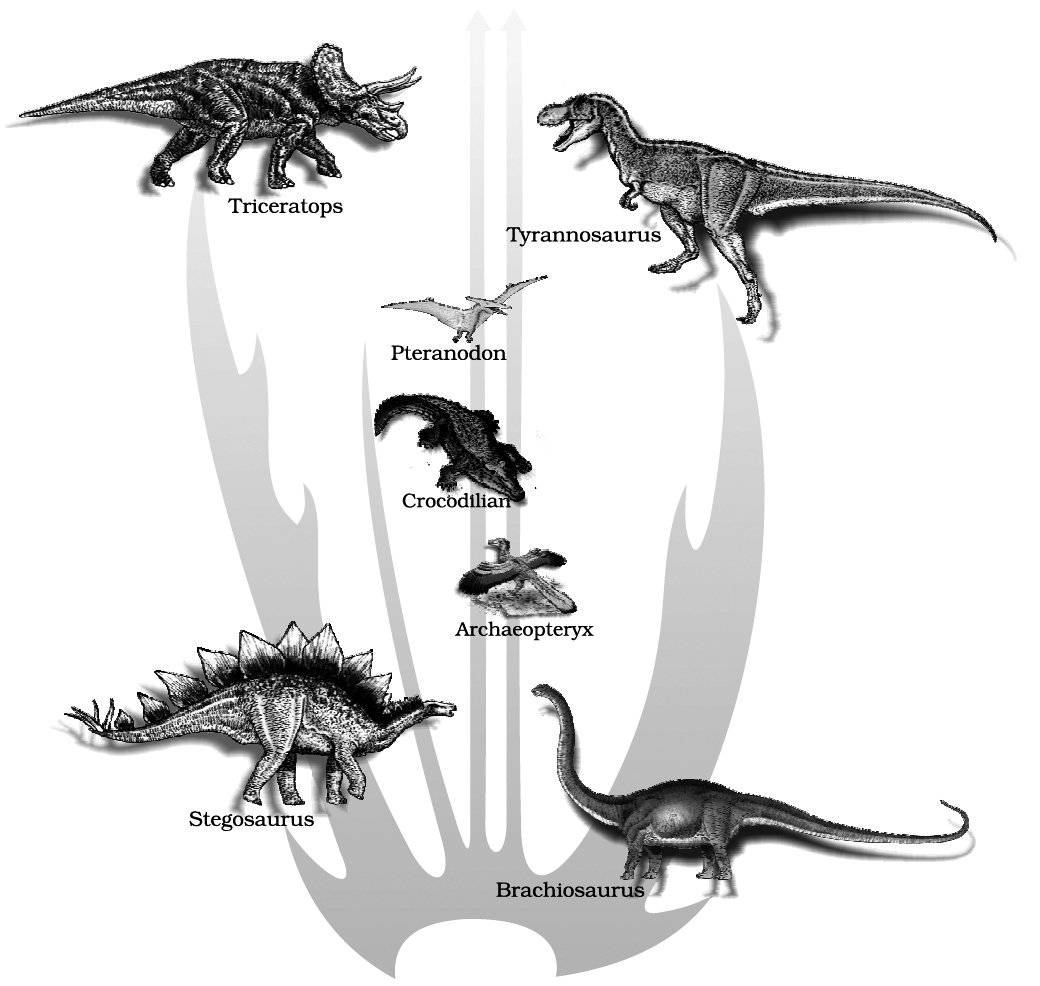
A family tree of dinosaurs and their living
modern day counterpart organisms like crocodiles and birds
The truth may live in between.
Small sized reptiles of that era still exist today.
The first mammals were like shrews.
Their fossils are small sized.
Mammals were viviparous and protected their unborn young inside the mother's body.
Mammals were more intelligent in sensing and avoiding danger at least.
When reptiles came down mammals took over this earth.
There were in South America mammals resembling horse, hippopotamus, bear, rabbit, etc.
Due to continental drift, when South America joined North America, these animals were over ridden by North American fauna.
Due to the same continental drift, pouched mammals of Australia survived because of lack of competition from any other mammal.
Lets we forget, some mammals live wholly in water.
Whales, dolphins, seals and sea cows are some examples.
Evolution of horse, elephant, dog, etc., are special stories of evolution.
You will learn about these in higher classes.
The most successful story is the evolution of man with language skills and self-consciousness.
THEORIES OF EVOLUTION
1. LAMARCK'S THEORY OF EVOLUTION
His theory is often called as the Theory of Inheritance of Acquired Characters or the Theory of Use and Disuse of Organ.
The first attempt to explain origin of species and their adaptation to the environment was done by Jean Baptiste Lamarck (1744-1829).
He was the greatest French naturalist.
Lamarck's theory was published in 1809 (year of Darwin 's birth) in his book 'Philosophie Zoologique'.
According to this theory the organisms undergo changes to adapt themselves to the environment.
The changes acquired by the organisms during their life time are passed on to the next generation.
He took the example of long neck of Giraffe, they continuously stretched their neck to reach to the vegetation on trees.
This acquired change was passed to the next generation.
He also gave the principle of Use and Disuse.
Use of an organ leads to strengthening of the organ, and disuse will lead to weakening of the organ.
Lamarck arranged his theory in the form of four postulates.
(i) Internal forces tend to increase size of the body.
(ii) Formation of new organs is the result of the need or want continuously felt by organisms Doctrine of Appetency/Desires.
(iii) Development and power of action of an organ is directly proportional to its use.
(iv) All changes acquired by the organism during its life are transmitted to the offsprings by the process of inheritance.
Concept Builder
Lamarck's theory was discarded by A. Weismann who gave the Theory of Germplasm.
He cut the tail of new born mice generation after generation but could not get tailless mice nor the mice developed shorter and shorter tail.
Today, again the faith in Lamarck's theory has been revived as it is said that if the environment influences the genes of the organisms, the acquired change will be transmitted to the next generation.
2. DARWIN'S THEORY OF EVOLUTION
Charles Robert Darwin put forward the concept of natural selection as the mechanism of evolution.
The theory was put forward along with Alfred Russell Wallace.
Darwin had written the book 'Origin of Species'.
Darwin was greatly influenced by 'An Essay on Population' written by Thomas Rev Malthus and he was also influenced by Charles Lyell's essays on "Principles of Geology".
Darwin was a British naturalist.
In 1831, at the age of 22, he was appointed upon a world survey ship of British government, H.M.S. Beagle.
For five years on his ship, Darwin explored the fauna and flora of continents and islands.
Branching descent and Natural Selection are the two key concepts of Darwinian Theory of Evolution.
According to Wallace's Chart, the main points of Darwin's theory of Natural Selection were as follows:
(i) High rate of reproduction
(ii) Total number almost constant
(iii) Struggle for existence
(iv) Variations
(v) Survival of fittest
(vi) Natural selection
All the successful organisms have a high Biotic Potential or Reproductive Rate.
The organisms produce a large number of offsprings that can possibly survive, example a mice produces a dozen of mice at one time.
A rabbit produces 6 young ones in a litter and there are four litter in a year.
A rabbit starts reproducing at the age of six months.
1. Not all but only some individuals which survive, reach adulthood, and those which reach adulthood, reproduce at different rates, this is called 'Differential Reproduction'.
2. The success in survival and reproduction depends upon the characteristic traits of an organism, example only those rabbits will survive which are fastest. There is 'Struggle for Existence' and in this there will be 'Survival of Fittest'. The Phrase 'Survival of Fittest' was first used by Herbert Spencer. The same context was asserted by Darwin as 'Natural Selection'.
So, evolution is the change in the genetic composition of the population which is brought about by natural selection which acts upon the variability in population.
Causes of Variations
1. Mutation is the ultimate source of variations.
2. At the next level is recombination .
3. Intermingling of two widely separated populations.
Weakness of Darwinism
He was not able to explain the cause of discontinuous variations observed by himself in nature and the mode of transmission of variants to the next generation.
In 1868, Darwin put forward the Theory of Pangenesis.
According to this theory, every organ of the body produces minute hereditary particles, called Pangenes or Gemmules and they are carried through the blood into the gametes.
Weismann's 'Theory of Germplasm' (1892) rejected Darwin's theory of pangenesis.
He established that the germ (sex), cells are set apart from other body (somatic) cells early in the embryonic development, so, only the changes in the germplasm affect the characteristics of future generations.
Alfred Wallace had written the book 'On the Tendencies of Varieties to Depart Indefinitely from the Original Type'.
Alfred Wallace (1823-1923), a naturalist from Dutch East Indies, was working on Malay Archipelago (present Indonesia).
Concept Builder
Erasmus Darwin :
Charles Darwin's grandfather, Erasmus Darwin, wrote about evolution more than 60 years before his grandson's theory was presented.
Erasmus Darwin cited things such as the metamorphosis of insects, the new varieties produced by selective breeding, the variations among similar organisms in different climates, and the similarities of vertebrate structure as evidence that all life was "produced from a similar living filament".
3. MUTATION THEORY
In 1901, Hugo de Vries proposed the Mutation Theory on the basis of his observation on the wild variety of evening primrose Oenothera lamarckiana.
According to mutation theory, new species originate as a result of large, discontinuous variations which appear suddenly.
The main features of mutation theory are as follows :
1. Mutations arise from time to time amongst the individuals of a naturally breeding population.
2. Mutations are heritable and establish new forms or species.
3. Mutations are large and sudden and are totally different from fluctuating variations of Darwin, which are small and directional.
4. Mutations may occur in any direction.
Human evolution
- Books Name
- A TEXT OF BIOLOGY - CLASS XII
- Publication
- ACME SMART PUBLICATION
- Course
- CBSE Class 12
- Subject
- Biology
HUMAN EVOLUTION
Place or origin of man :
It has been established that Dryopithecus is one of the oldest fossil which in turn evolved into apes and men.
The origin and evolution of man can be studied in the following three major headings : prior to ape men, ape men including prehistoric man and true men including the living modern man.
A. Prior to Ape Men
1. Dryopithecus. Discovery:
The fossil of Dryopithecus africanus was discovered from Miocene rocks of Africa and Europe.
It lived about 15 million years ago.
Dryopithecus and Ramapithecus were hairy and walked like gorillas and chimpanzees.
Ramaplthecus was more man like while Dryoplthecus was more ape like.
Dryopithecus is the direct ancestor of modern day apes.
Characteristics :
It was ape-like, but had arms and legs of the same length.
Heels in its feet indicate its semierect posture.
It had large brain, a large muzzle and large canines.
It was without brow ridges.
It was arboreal, knuckle-walker and ate soft fruits and leaves.
Dryopithecus africanus is regarded a common ancestor of man and apes (gibbons, orangutan, chimpanzee and gorilla).
It is also called proconsul.
2. Proconsul. Discovery:
Proconsul africanus or o. africanus was discovered by Louis S.B. Leakey in 1948 from the rocks around lake Victoria of Kenya, Africa.
It lived in early Miocene epoch.
Characteristics :
It was morphologically intermediate between apes and man in many features.
It had rounded man-like forehead and long, pointed ape like canines.
It moved upon land on all the four limbs and hence is not considered amongst the direct ancestors of man.
Proconsul gave rise to the ancestors of chimpanzee and gorilla in the Pliocene, about 4 million years ago.
The chimpanzee and gorilla diverged from each other only about 2.3 million years ago, in the Pleistocene epoch.
3. Sivapithecus. Discovery :
This fossil was discovered from middle and late Pliocene rocks of Shivalik Hills of India, hence it is named Shivapithecus.
Characteristics :
It was like Dryopithecus. Its fore limbs, skull and brain were like those of monkeys, while the face, jaws and teeth resembled those of apes.
4. Ramapithecus. Discovery:
It has been established that in late Miocene epoch, Dryopithecus gave rise to Ramapithecus ('Rama' =The hero of Indian legend, Pithecus =Ape) which was on the direct line of human evolution.
Ramapithecus survived from late Miocene to Pliocene.
Thus, he appeared about 14-15 million years ago.
Fossil of Ramapithecus was discovered by Edward Lewis (1932) from Pliocene rocks of Shivalik Hills of India.
Kenyapithecus wickeri was discovered by L.S.B. Leakey (1962) from Pliocene rocks of Kenya (Africa).
It was similar to Ramapithecus. But Ramapithecus was older than Kenyapithecus.
B. Ape-men Including Prehistoric Men
1. Australopithecus (First Ape-man) :
The early human stock gave rise to Australopithecus.
It is the connecting link between apes and man.
Discovery. Raymond Dart (1924), South African anthropologist, discovered the fossil of Australopithecus africanus (African Ape-man) from Pliocene rocks near Tuang in Africa.
They appeared about 5 million years ago.
Actually skull discovered by Dart was of 5-6 year old baby so it is also called "Tuang baby".
Some fossils of A. africanuswere also discovered from Pleistocene epoch.
Two mya, Australopithecines probably lived in east African grasslands.
Evidence shows they hunted with stone weapons but essentially ate fruits.
Characteristics :
Australopithecus africanus was about 1.5 metres high and had human as well as ape characters.
It was with bipedal locomotion, omnivorous diet and had erect posture.
It had human like teeth, but it had more of an ape brain than a human brain.
Its brain capacity was about 500 cc., similar to that of an ape.
He lived in caves.
Brow ridges projected over the eyes.
It did not have chin.
There was lumbar curve in the vertebral column.
The pelvis was broad. Australopithecus africanus existed until about 1.5 million years ago and gave rise to Homo habilis, about two million years ago.
Australopithecus africanus also gave rise to man-like apes called Australopithecus robustus and Australopithecus boisei along a separate line that ended blindly (They did not give rise to any other creatures).
In 1981 Donald Johanson, found a 3.2 million years old skeleton of a female human ancestor.
He nicknamed it Lucy.
Lucy's scientific name is Australopithecus afarensis.
Six species of Australopithecus are known.
These are A. africanus (African Ape man, Southern Ape or Tuang baby), A. afarensis, (Lucy), A. ramidus, A. aethiopicus, A. robustus and A. boisei. So we can say that Australopithecus had two main types.
(i) Gracile type: Australopithecus afarensis (Johanson) represented by fossil Lucy with small brain, small molar teeth pelvic girdles and short fingers like human.
(ii) Robust type: A. robustus had heavier body structure and massive check tooth (also called originally Paranthropus) cranial capacity -600cm2.
(Other examples -Zinjanthropus / A. boisei of R. Leakey -Africa, Meganthropus from -Java)

Skull and reconstructed head
2. Homo habilis (Able or Skillful man, The tool maker, or 'Handy man'). Discovery:
Louis S.B. Leakey and his wife Mary Leakey (1960) discovered the fossils of Homo habifis from Pleistocene rocks of Olduvai Gorge in East Africa.
He lived in Africa, about 2 million years ago.
The first human like being the hominid was Homo habilis. They probably did not eat meat.
Characteristics:
He was about 1.2 to 1.5 metres tall.
He had bipedal locomotion and moved erect.
It had about 650-800 cc. cranial capacity.
The teeth were like that of modern man.
Homo habilis (habilis = mentally able or skillful) was the first tool maker and used tools of chipped stones extensively.
It is also called handy man because heaps of tools found with these fossils included sharpened stones which indicate that Homo habilis was capable of 'making tools'.
He also led community life in caves and greatly cared for the young ones.
3. Homo erectus (Erect man) :
Homo erectus appeared about 1.5 million years ago, in middle Pleistocene.
Homo erectus probably ate meat.
He is called middle pleistocene man. H. erectus evolved from Homo habilis.
He was about 1.5-1.8 metres tall.
Homo erectus males were probably larger than females. He had erect posture.
His skull was flatter than that of modern man.
He had protruding jaws, projecting brow ridges and small canines and large molar teeth.
The cranial capacity was 900 cc.
Cranium was domed to accommodate the large brain.
He was omnivorous.
He made more elaborate tools of stones and bones, hunted big game and perhaps knew the use of fire.
Skull and reconstructed head
Homo erectus includes three fossils : Java Ape-man, Peking man and Heidelberg man.
(a) Java Ape-man. Discovery:
In 1891, Eugene Dubois discovered a fossil from Pleistocene rocks in central Java (Island of Indonesia).
Eugene Dubois named it as Pithecanthropus erectus.
Pithecanthropus means 'ape man' Mayer in 1950 assigned it as Homo erectus erectus.
Characteristics :
Body 1.65 to 1.75 metres tall and weight about 70 kg.
Legs long and erect, but body slightly bent when moving.
Inconspicuous chin and somewhat broader nose.
Forehead low and receding, but brow ridges high, like those of apes.
Skull cap thick and heavy, flattened infront.
Cranial capacity 800 to 1000 ce. (average 950 cc).
Lower jaw large and heavy.
Teeth large, but quite like those of modem man, except larger canines of the lower jaw.
Lips thick and protruding.
He was omnivorous and cannibal.
Perhaps, he was the first prehistoric man to make use of fire for hunting, defence and cooking.
(b) Peking Man. Discovery:
W.C. Pei (1924) discovered the fossils of Peking man from the lime stone caves of Choukoutein near Peking (Beijing -capital of China was formerly known as Peking) and named them Sinanthropus.
Davidson Black (1927) named it Sinanthropus pekinensis.
Mayer (1950) renamed it as Homo erectus pekinensis (a subspecies).
The Pleistocene rocks from which fossils of Peking man were excavated are about 6 lakh years old.
Characteristics :
Placing Java ape man and Peking man as subspecies of Homo erectus has a sound basis, because of close similarities between these.
The body structure was quite similar in both.
Being about 1.55 to 1.60 metres tall, Peking man was slightly shorter and a little lighter and weaker.
The only noticeable difference of Peking man from Java ape man was its large cranial capacity, which ranged from 850 to 1100 cc.
Like Java ape man, the Peking man was omnivorous and cannibal.
There is a clear evidence of use of fire by it.
It has been confirmed that both Java and Peking men used to live in caves in small groups or tribes.
The tools of Peking man were relatively more sophisticated.
(c) Heidelberg man. Discovery :
In 1908 one of the most perfect fossil jaws belonging to middle Pleistocene was found by workmen working near Heidelberg, Germany.
It was shown to Otto Schoetensack, who gets the credit for its discovery. It was named Homo erectus heidelbergensis.
Characteristics :
It had lower jaw with all the teeth.
The teeth were human like.
The massive jaw was ape-like.
He used the tools and fire.
The cranial capacity is believed to be about 1300 cc; which is intermediate between those of erect man (H. erectus) and Neanderthal man (H. sapiens neanderthalensis).
Thus, it is regarded as intermediate between pithecanthropines and Neanderthals.
C. True Men Including the Living Modern Man
1. Neanderthal Man (Homo sapiens neanderthalensis). Discovery:
Fossils of Neanderthal man were first obtained from Neanderthal Valley in Germany from the late Pleistocene epoch by C. Fuhlrott (1856).
Later, many other fossils were excavated in various countries by different palaeontologists.

Skull and reconstructed head
Characteristics:
He had slightly prognathous face.
Neanderthal man walked upright, as we do, and had low brows, receding jaws, and high domed heads.
If there was anything truly different about them, it was that they were much stockier than we are.
The cranial capacity was 1,300 to 1,600 cubic centimetres.
Neanderthal man existed half a million years ago, but was most numerous from about 100,000 years ago.
Became extinct 30,000 years ago.
Neanderthal man was the legendary cave dweller, having heavy brow ridge and humped back.
He was adapted to a cold environment, who encountered a succession of glaciers that passed over most of the northern temperate regions of the world.
He was not only skilled hunter but true predator, a specialization that did not occur among hominids before or after them.
The Neanderthal man was cannibal and fashioned the skin into clothing to protect himself against the harsh climate.
Natural caves became campsites that were illuminated and heated by fire.
It is believed that he buried his dead with flowers and tools. He may have had a religion.
It is usually considered that Homo sapiens neanderthalensis did not evolve into Homo sapiens.
2. Cro-magnon man (Homo sapiens tossilis). Discovery:
It has been known as Cro-Magnon man, because its fossils were first discovered in 1868 from Cro-Magnon rocks of France by MacGregor.
Cro-Magnon man emerged about 34,000 years ago in Holocene epoch.
Thus, he is regarded as most recent ancestor of today's man.
Characteristics:
The Cro-Magnon man had, like us, about 1.8 metres tall, well-built body.
His face was perfectly orthognathous with an arrow, elevated nose, braad and arched forehead, moderate brow-ridges, strong jaws with man-like dentition, and a well developed chin.
His cranial capacity was, however, somewhat more than ours, being about 1650 cc.
It is, therefore, believed that Cro-Magnon man was somewhat more intelligent and cultured than the man of today.
It could walk and run faster and lived with families in caves.
It made excellent tools and even ornaments, not only of stones and bones, but also of elephant tusks.
Its tools included spears, bows and arrows, as he was omnivorous.
Use of the skin clothes by this man is also confirmed.
A number of cave paintings done by Cro-Magnon man have been discovered.
The Cro-Magnon man was the direct ancestor of the living modern man.
Prehistoric cave art developed about 18,000 years ago.
3. The Living Modern Man (Homo sapiens sapiens). Discovery :
Further evolution of man after Cro-Magnon involves the evolution of culture rather than that of anatomy.
Homo sapiens sapiens appeared about 25,000 years ago in Holocene epoch and started spreading all over the world about 10,000 years ago.
Agriculture came around 10,000 years back and human settlements started.
MODERN HUMANS
Homo sapiens
The evolutionary journey to modern humans ends with the appearance, about five hundred thousand years ago, of Homo sapiens ("wise man"), our own species.
We are newcomers to the human family -H. sapiens has not been around nearly as long as H. erectus was.
Still, humans have changed quite a bit, since those first days.
Concept Builder
Out of Africa -Again
The origin of human races is a much-debated point among scientists studying human evolution.
Many argue that the different races evolved from H. erectus independently, and that each adapted to a different place -Orientals in Asia, Caucasians in Europe, Aborigines in Australia, and so on.
Others believe that the same species would be unlikely to evolve more than once and argue that human races appeared after H. sapiens evolved from H. erectus.
Recently, scientists studying mitochondrial DNA from living humans all over the world have argued that their research shows that all human races originated from one H. sapiens ancestor in Africa.
Scientists looked at mitochondrial DNA to study evolution, because the DNA within mitochondria is transmitted only by females.
Female's eggs carry many mitochondria within them that become part of a new baby, while sperms contribute no mitochondria to the new baby.
Sperms carry their mitochondria wrapped around their tails and so do not inject them into the egg during fertilization.
For that reason, particular versions of a mitochondrial gene can be traced back through a family tree, from mother to grandmother to great-grandmother.
Human races evolved only recently in the evolutionary scale of things, and there has not been enough time for many mitochondrial DNA differences to accumulate, so the exact human tree cannot be reliably traced using this approach.
So far, however, the greatest number of different mitochondrial DNA sequences occur among modern Africans.
Since DNA accumulates mutations over time, the oldest DNA should show the largest number of mutations.
This result thus argues, that humans have been living in Africa longer than on any other continent.
While researchers are not in complete consensus, this line of investigation appears to suggest that H. sapiens evolved in Africa and that the human races evolved after that, and not independently from separate species of H. erectus.
If this Is correct, then H. sapiens was born in Africa and from there spread to all parts of the world, retracing the path taken by H. erectus half a million years before.
baby chimpanzee and adult chimpanzee. The skull of baby chimpanzee is
more like adult human skull than adult chimpanzee skull
Homology In Chromosomes of Man and Great Apes
The somatic cells of humans contain 46 chromosomes (44 autosomes and 2 sex- chromosomes).
Human chromosomes are usually obtained by culturing certain types of white blood cells from the peripheral blood.
They can then be treated with specific stains to produce characteristic bands along the length of each chromosome.
The pattern of banding so obtained is unique for each pair of chromosomes.
Banding techniques enable the identification of individual chromosomes and their parts.
The diploid number of chromosomes in gorilla, chimpanzee and orangutan is 48.
Comparisons have been made between banded chromosomes of man and those of the great apes.
The total amount of DNA in human diploid cells and that of the great apes are not dissimilar.
But what is most interesting from an evolutionary viewpoint, is that the banding pattern of individual human chromosomes is very similar and in some instances, identical to the banding pattern of apparently homologous chromosomes in the great apes.
Diagrammatic representations of the banding pattern of human chromosome numbers 3 and 6 are compared with those of particular autosomes in the chimpanzee.
This remarkable similarity in the fine structural organisation of the chromosomes is understandable only in terms of a common origin for man and chimpanzee.
Concept Builder
Cope's Law: It states that there is a tendency for animals to increase in size during the long course of evolution.
Bergman's Law : It states that warm blooded animals become larger in the northern and colder parts of their range.
Allen's Law : It states, that in animals which live in very cold climates, their extremities such as ears, tails etc. become progressively smaller.
Gause's Law: (Gause, 1934) or the Competitive exclusion Principle (Hardin, 1960). It states that two species having the same ecological requirements cannot continue to occupy indefinitely the same habitat.
Gloger's rule: It states that among warm blooded animals, those living in warm and moist climate develop more melanin pigment (are darker than animals in cold, dry climates) whereas forms in dry, hot climates have more yellow and red pigment.
Jordan's rule: Temperature also influences the morphology of certain fishes and is found to have some relation with the number of vertebrae. Fishes inhabiting water of low temperature tend to have more vertebrae than those of warmer water.
Some Important Points
1. Mars has CO2, water vapours and supposed to have life. CO2 is present in traces. It has no green house effect, hence it is very cold and does not support life. Mercury and moon do not have any sign of life due to the absence of water vapours. This extra terrestrial origin of life was visualised by Hoyle and Wickremsinghe.
2. Darwin used the term 'warm little pond' for early hot sea, rich in biomolecules. These primitive seas were alkaline.
3. K. Bahadur exposed ammonia, formaldehyde, and ferrous chloride to strong sunlight and obtained a mixture of amino acids called Jivam.
4. Variation In behaviour -Cicada insect has a life span of 17 years and it emerges from soil, remains alive for 5 weeks and then dies after mating and laying eggs. Dolphins can imitate and laugh. Bat can detect small insects of size (o.oa mm). Male baya (weaver's bird) of India builds its elaborate nest and decorates it with colourful petals to attract female.
5. Multiformity among organisms -Internal differentiation increases with the progress in evolution. Human beings are one of the most recently evolved animals. They show the following features :
(a) Total length of blood vessels in our body is 96,000 km.
(b) Fastest nerve impulse travels at the rate of 532 km per hour.
(c) Internal area of our lungs is 93-100 m2 which is 40 times than the external surface area of our body.
(d) Human brains have 10,000 million nerve cells.
(e) We have more body hair than apes but shorter and softer.
(f) O2 disappears from atmosphere at 16 km height.
(g) We remain blind for 30 minutes per day by blinking our eyes.
(h) Bones are as strong as concrete and as hard as granite but far lighter than both.
(i) We retain only 18% of total that we learnt yesterday.
6. Synapsid reptiles were mammal like reptiles that gave rise to mammals. They had a single temporal fossa on the lateral side of skull and heterodont teeth. They are extinct. They originated in Permian period.
7. In 1858 Dr. P.L. Sclater divided for first time the earth into six regions (realms) according to the distribution of birds. Later on Alfred Russel Wallace (1876) classified the earth into six regions (realms) for all animals and plants.
8. In all animals, early development is similar i.e., passes through morula blastula gastrula stages, showing their common origin.
9. Early embryos of all the vertebrates show basic similarity in having somites, tail, gill clefts, notochord etc. These traits can be explained as a character of evolution.
10. Any vertebrate organ also passes through different stages during development. e.g., mammalian heart is initially two chambered, then becomes three chambered and then becomes four chambered.
Development of all triploblastic animals starts from zygote, undergoes similar changes to form gastrula having 3 primary germ layers (ectoderm, mesoderm and endoderm) which have same fate in organogenesis. Early embryos of different vertebrates resemble in possessing similar structures like gill slits, notochord, tail etc. Not only this, in the course of development, at different stages, an embryo looks like the embryo of different phyla form, which the given organism has evolved.
It can be explained on the basis of Recapitulation theory (Von Baer) / Biogenetic law (Haeckel) which states that "ontogeny (developmental history of an individual) repeats phylogeny" (developmental history of race).
11. Types of fossils
(i) Macrofossils: Larger than 1 cm in size.
(ii) Unusual fossils: Sudden preservation of entire organism e.g., Solenhofen Limestone quarry of Southern Germany -containing fossils of Archaeopteryx.
(iii) Bioclast: Fossils or fragments of fossils enclosed in sediments. The term is usually applied to thin sections of fossils under microscope.
(iv) Gastroliths: These are found in abundance in the body cavities of certain reptiles.
(v) Moulds and casts: The material surrounding the fossil hardens and preserves the outer details. The actual bodies diSintegrate and are removed by slippage of the ground leaving the harden cavities called moulds. When moulds are filled with natural deposits, they are·called as casts e.g., fossils of Pompeii city buried in volcanic ash of mount Vesuvious in A.D. 79.
(vi) Tracks and trails : The footprints or tracks left in the soft moist mud gets hardened up e.g., tracks of amphibians discovered near Pittsburg, Germany from Pennsylvanian period.
12. Preservation in ice : Woolly mammoths from Siberia. The flesh is so well preserved that it can be fed to dogs. Discovered from Lena delta in 1790 and Siberia in 1901.
13. Fossils in petroleum springs and asphalts : Rancho La Brea now in Los Angeles.
14. Fossils in resins and ambers: Fossil fly in amber from Baltic forests of Europe during Oligocene period.
15. The process of fossilization to preserve the finer details is known as histometabasis.
16. Mummies: Bodies of dead animals or plants become dehydrated in the deserts and are preserved as mummies.
17. T. Dobzhansky wrote the book 'Genetics and origin of species'.
18. Darlington wrote the book 'The evolution of genetic systems'.
19. Darwin wrote 'Descent of Man and Selection in relation of sex' in which he put forward his theory of evolution of man from ape like ancestors.
20. Law of Superposition : The lower strata of a geological formation was first to be deposited and is the oldest.
21. Willston's Rule : During evolution of lineage, serially homologue parts tend to reduce in number but get more and more differentiated e.g., prawn's leg.
22. Allometry: The study of differential growth rate was called allometry.
23. Missing links : The fossils which act as transition between two present day groups of organisms ego Archaeopteryx -a fossil of crow sized toothed bird act as a link between reptiles and birds.
24. Empedocales (493 -435 B.C.) -regarded as "Father of the Concept of Evolution".
25. Seymouria (Extinct Reptile) is a connecting link between amphibia and reptilia.
26. Lycaenops (Extinct Reptile) is a connecting link between reptiles and mammals.
27. Wallace's Line: In 1863 A.R. Wallace drew an imaginary dividing line on the map between the Oriental and Australian realms (regions). This line is known as Wallace's line.
28. Sibling species: Species which are morphologically looking similar but are reproductively isolated, are called sibling species.
29. Living Fossils: A living fossil is a living animal of ancient origin with many primitive characters. A living fossil has been living as such from the time of origin without many changes.
30. Eugenics: It is the branch of science which deals with improvement of human race genetically. It can also be divided into two types : Negative eugenics and Positive eugenics. Under negative eugenics, people with inferior and undesirable (dysgenic) traits are prevented from reproducing.
31.
32. Homo sapiens or modern man is a member of the order primate, sub-order Anthropoidea. Primates have supposed to have evolved from primitive, tiny, insect eating quadruped, similar to modern tree shrews, which lived between 75 and 60 MYA during Eocene period. These belong to order Insectivora. Two evolutionary lines diverged leading to present day prosimians (treeshrews, lemurs, loris and tarsier) and the Anthropoidea (including old world and new world monkey, Ape and man)
33. The first (Ape + Man) ancestor originated in Oligocene period 30-35 MY A under the name Propliopithecus. (Fayiim deposits of Egypt). It is represented by fossil jaws and teeth. Aegyptopithecus is contemporary of Prppliopithecus (Kahira).
34. Dryopithecus -The Oligocene ancestor gave rise to Miocene group of (Apes + Man) called Dryopithecus (formerly known as proconsul). It lived in Africa and Asia. It had semi-erect posture with hind limbs and fore limbs of the same size. Hands and skull were monkey like, fore head like humans, jaw and dentition like apes. Shivapithecus discovered by Chopra Simon team is another fossil ape from Shivalik hills in India (derived from Dryopithecian stock).
An aberrant branch from Oryopithecus evolved in late Miocene early Pliocene, Oreopithecus which later on became extinct. Ramapithecus (= kenyapithecus) 14-15 MYA few teeth and fragments of jaw. Believed to have evolved from Dryopithecus in Shivalik hills in India during late Miocene and early Pliocene.
35. Gremaldi -From caves in a village Grimaldi on the Mediterranean coast, cranial capacity 1655 cc., believed to have given rise to Negroid stock.
Chancelade -Rock shelter near chancelede in Dordogne France, Cranial capacity -1450 cc., gave rise to Modern Eskimos.
36. Modern Man -H.sapiens sapiens evolved about 25,000 years ago but spread to various parts of world about 10000 -11000 years ago. There is thining of skull bones, slight reduction in cranial capacity (1400 -1450 cm3), four flexors in vertebral column and slight raising of skull cap. Modern man underwent cultural evolution. Paleolithic (age of tools of stones, bones, cave paintings). Mesolithic (age of animal husbandry, development of language reading and writings). Neolithic (development of agriculture, manufacture of pottery and clothes). Bronze age followed by iron age.
Forgery of Piltdown Man (Eoanthropus dawsoni) Charles Dawson in 1921 reconstructed the skull from the cranium of modern man and lower jaw of ape. The fossil skull known as Piltdown, the English hamlet where it was found.
37.

38. Transitional forms connecting Home erectus with Homo sapiens have been uncovered from Europe -Steinheim skull (Germany) Swanscombe skull (2nd Interglacial period) Fontechvade skulls (France 3rd Interglacial period) Ehringsdore skull. The above all are called as early H.sapiens.
The course of evolution of man started in Africa.

 ACME SMART PUBLICATION
ACME SMART PUBLICATION
